Last active
December 22, 2020 17:44
-
-
Save ali-ramadhan/1edceaa78e550ca220e591d5c6db9aed to your computer and use it in GitHub Desktop.
Ensemble Kalman inversion example by @sandreza using CalibrateEmulateSample.jl
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| using Distributions | |
| using LinearAlgebra | |
| using Random | |
| using Test | |
| using Plots | |
| using CalibrateEmulateSample.EKP | |
| using CalibrateEmulateSample.ParameterDistributionStorage | |
| ## Seed for pseudo-random number generator | |
| rng_seed = 41 | |
| Random.seed!(rng_seed) | |
| ## Generate data from a linear model: a regression problem with `n_par` parameters | |
| ## and `n_obs` observations: G(u) = A×u, where A : R^n_par -> R | |
| n_obs = 10 # Number of synthetic observations from G(u) | |
| n_par = 2 # Number of parameteres | |
| u_star = [-1, 2] # True parameters | |
| noise_level = 0.1 # Defining the observation noise level | |
| Γy = noise_level * Matrix(I, n_obs, n_obs) # Independent noise for synthetic observations | |
| noise = MvNormal(zeros(n_obs), Γy) | |
| C = [1 -0.9; -0.9 1] # Correlation structure for linear operator | |
| A = rand(MvNormal(zeros(2,), C), n_obs)' # Linear operator in R^{n_obs × n_par} | |
| @test size(A) == (n_obs, n_par) | |
| y_star = A * u_star | |
| y_obs = y_star + 0 * rand(noise) | |
| @test size(y_star) == (n_obs,) | |
| ## Define linear model | |
| G(u) = A * u | |
| @test norm(y_star - G(u_star)) < n_obs * noise_level^2 | |
| ## Define prior information on parameters | |
| prior_distns = [Parameterized(Normal(1.0, √2)), | |
| Parameterized(Normal(1.0, √2))] | |
| constraints = [[no_constraint()], [no_constraint()]] | |
| prior_names = ["u1", "u2"] | |
| prior = ParameterDistribution(prior_distns, constraints, prior_names) | |
| prior_mean = reshape(get_mean(prior),:) | |
| prior_cov = get_cov(prior) # Assuming independence of u1 and u2 | |
| ## Calibrate (1): Ensemble Kalman Inversion | |
| N_ens = 50 # number of ensemble members | |
| N_iter = 20 # number of EKI iterations | |
| initial_ensemble = EKP.construct_initial_ensemble(prior,N_ens, rng_seed=rng_seed) | |
| @test size(initial_ensemble) == (N_ens, n_par) | |
| ekiobj = EKP.EKObj(initial_ensemble, y_obs, Γy, Inversion()) | |
| ## EKI iterations | |
| for i in 1:N_iter | |
| params_i = ekiobj.u[end] | |
| g_ens = G(params_i')' | |
| EKP.update_ensemble!(ekiobj, g_ens) | |
| end | |
| ## EKI results: Test if ensemble has collapsed toward the true parameter values | |
| eki_final_result = vec(mean(ekiobj.u[end], dims=1)) | |
| ## Plot results | |
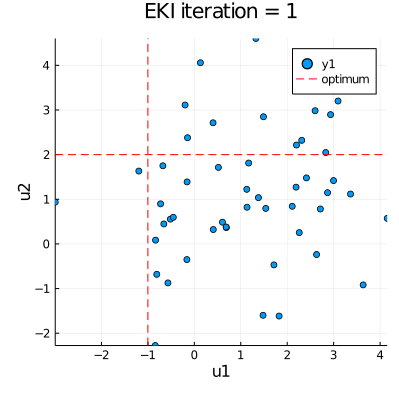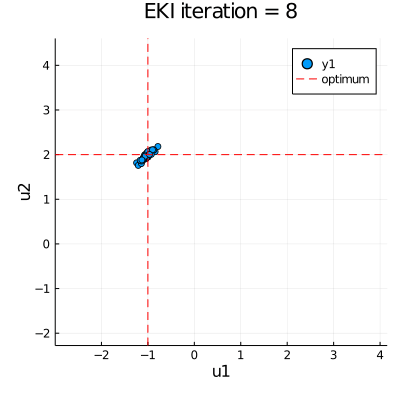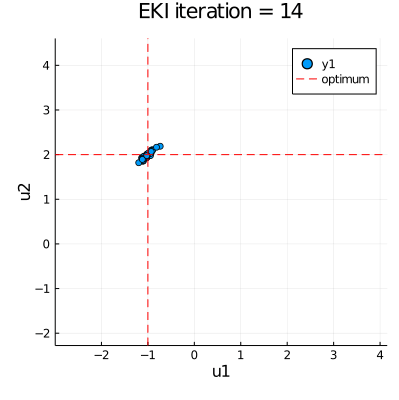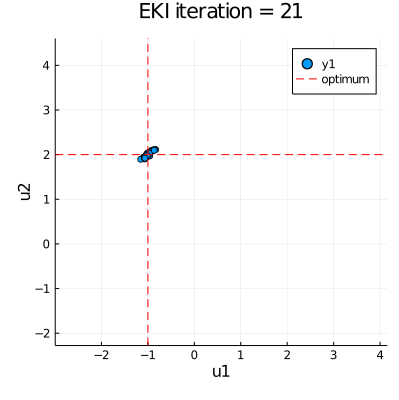
| anim = @animate for i in eachindex(ekiobj.u) | |
| p = plot(ekiobj.u[i][:,1], ekiobj.u[i][:,2], seriestype=:scatter, | |
| xlims=extrema(ekiobj.u[1][:,1]), ylims=extrema(ekiobj.u[1][:,2])) | |
| plot!([u_star[1]], xaxis="u1", yaxis="u2", seriestype="vline", | |
| linestyle=:dash, linecolor=:red, label=false, title="EKI iteration = " * string(i)) | |
| plot!([u_star[2]], seriestype="hline", linestyle=:dash, linecolor=:red, label="optimum") | |
| end | |
| gif(anim, "EKI.gif", fps = 30) |
Author
ali-ramadhan
commented
Dec 22, 2020
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment