library(brms)
library(bayestestR)
m <- brm(
mpg ~ 1,
family = lognormal(),
data = mtcars,
backend = "cmdstanr"
)
m_pars <- as.data.frame(m)
mu <- m_pars$b_Intercept
sigma <- m_pars$sigma
nd <- data.frame(whatever = NA)linpred gives mu (log(median)), as expected:
posterior_linpred(m, newdata = nd) |> as.data.frame() |>
cbind(Median = mu) |>
exp() |>
describe_posterior()
#> Summary of Posterior Distribution
#>
#> Parameter | Median | 95% CI | pd | ROPE | % in ROPE
#> ----------------------------------------------------------------------
#> V1 | 19.26 | [17.29, 21.40] | 100% | [-0.10, 0.10] | 0%
#> Median | 19.26 | [17.29, 21.40] | 100% | [-0.10, 0.10] | 0%
But the epred knows how to get the actual expected values (exp(mu + (sigma^2)/2)):
posterior_epred(m, newdata = nd) |> as.data.frame() |>
cbind(Manual = exp(mu + (sigma^2)/2)) |>
describe_posterior()
#> Summary of Posterior Distribution
#>
#> Parameter | Median | 95% CI | pd | ROPE | % in ROPE
#> ----------------------------------------------------------------------
#> V1 | 20.21 | [18.03, 22.56] | 100% | [-0.10, 0.10] | 0%
#> Manual | 20.21 | [18.03, 22.56] | 100% | [-0.10, 0.10] | 0%In other words, brms provides here more than just a syntactic-sugar-log-transformation.
In this one, the mean of Y (not log(Y)) is modeled.
lognormal2 <- custom_family(
"lognormal2",
dpars = c("mu", "sigma"),
links = c("identity", "log"),
lb = c(0, 0),
type = "real"
)
stan_funs <- "
real lognormal2_lpdf(real Y, real mu, real sigma) {
return lognormal_lpdf(Y | log(mu) - (sigma ^ 2)/2, sigma);
}
"
stanvars <- stanvar(scode = stan_funs, block = "functions")
posterior_epred_lognormal2 <- function(prep) {
brms::get_dpar(prep, "mu")
}m2 <- brm(
mpg ~ hp,
family = lognormal2,
data = mtcars,
backend = "cmdstanr",
stanvars = stanvars
)
ggpredict(m2, "hp") |> plot() # linear!library(ggplot2)
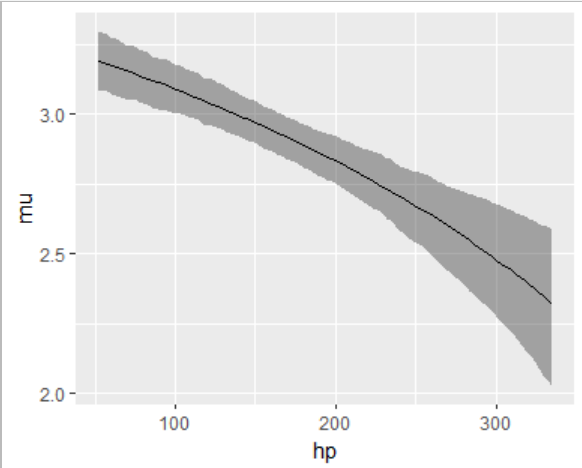
nd <- data.frame(hp = with(mtcars, seq(min(hp), max(hp), length = 100)))
mu_ <- posterior_linpred(m2, newdata = nd, dpar = "mu")
sigma <- posterior_linpred(m2, newdata = nd, dpar = "sigma")
true_mu <- log(mu_) - (sigma ^ 2)/2
true_mu |>
as.data.frame() |> describe_posterior( test = NULL) |>
cbind(nd) |>
ggplot(aes(hp, Median)) +
geom_ribbon(aes(ymin = CI_low, ymax = CI_high), alpha = 0.4) +
geom_line() +
labs(y = "mu")
Alt, instead of modeling the expected value, we can model
exp(mu)- the mu parameter on the response scale (a linear X to mu relationship).