Generating a graphviz dot file equivalent to the mol file of pred(3) in chemlambda v2: pred_3.mol (original name was erroneous)
The lambda calculus expression for pred(3) (predecessor(3) == 2) is PRED := λn.λf.λx.n (λg.λh.h (g f)) (λu.x) (λu.u) (ref wikipedia)
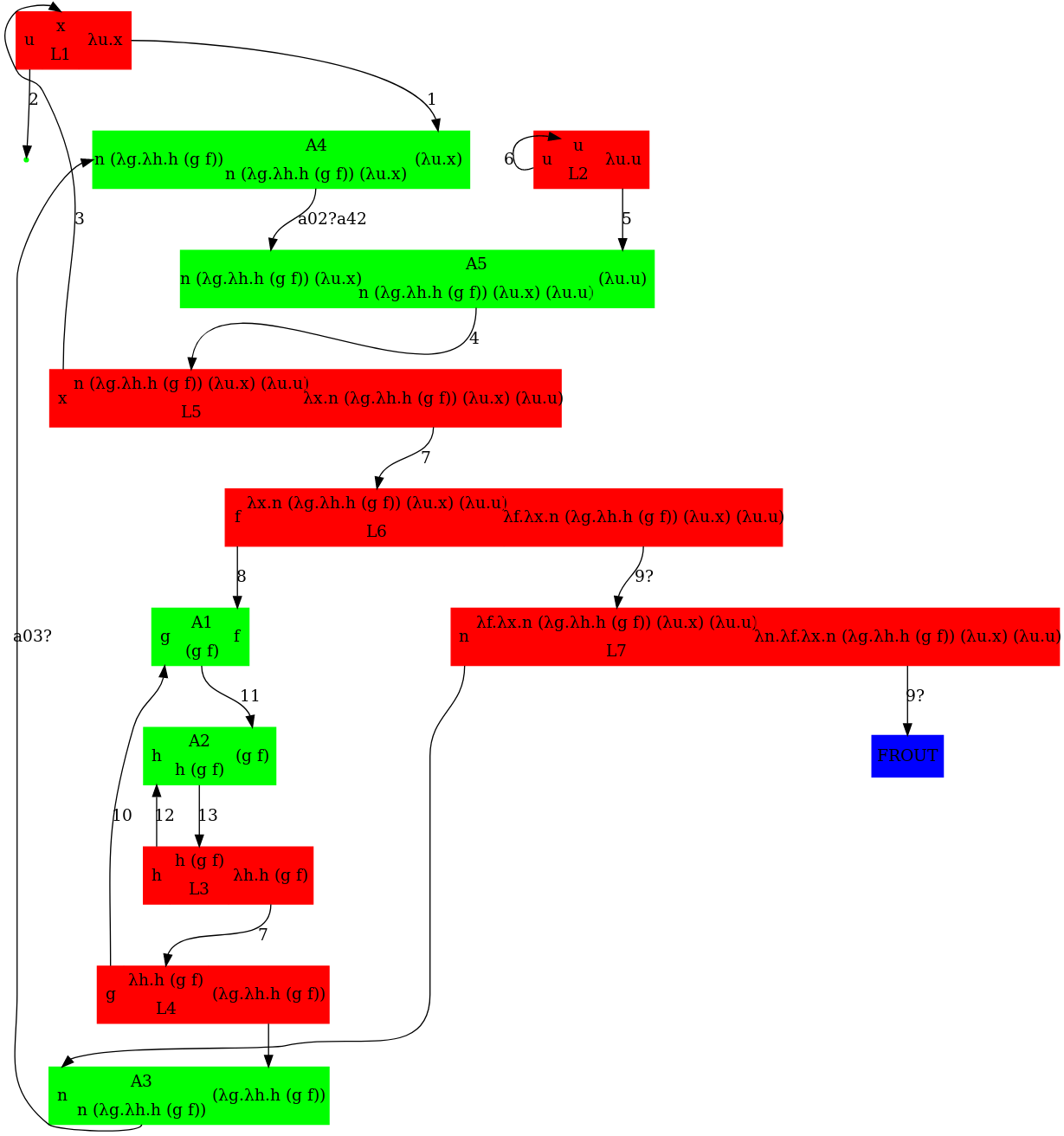
Writing the above expression as nodes in a graph leads to the below graph. The corresponding dot file was written manually and available further below in this gist. An annotated version of pred_3.mol to help compare it to pred_3.dot is available further below in this gist.
To generate the dot file automatically from lambda terms, check http://www.teamshadi.net/chemlambda-js/ (a fork from this jsfiddle ). Its current output for pred(3) matches with the manually specified graph below.
Version 4 (2019-04-11): shifted indeces back by 1 (e.g. L1 to L0) to match with the auto-generated graph from here
Compare the above graph to the chemlambda d3.js graph for pred_3.mol here
To experiment with graphviz, use
http://www.webgraphviz.com/ or
https://dreampuf.github.io/GraphvizOnline
http://viz-js.com/
Notes:
- This work was prompted from chamlambda issue #6
- Older versions of graphs:

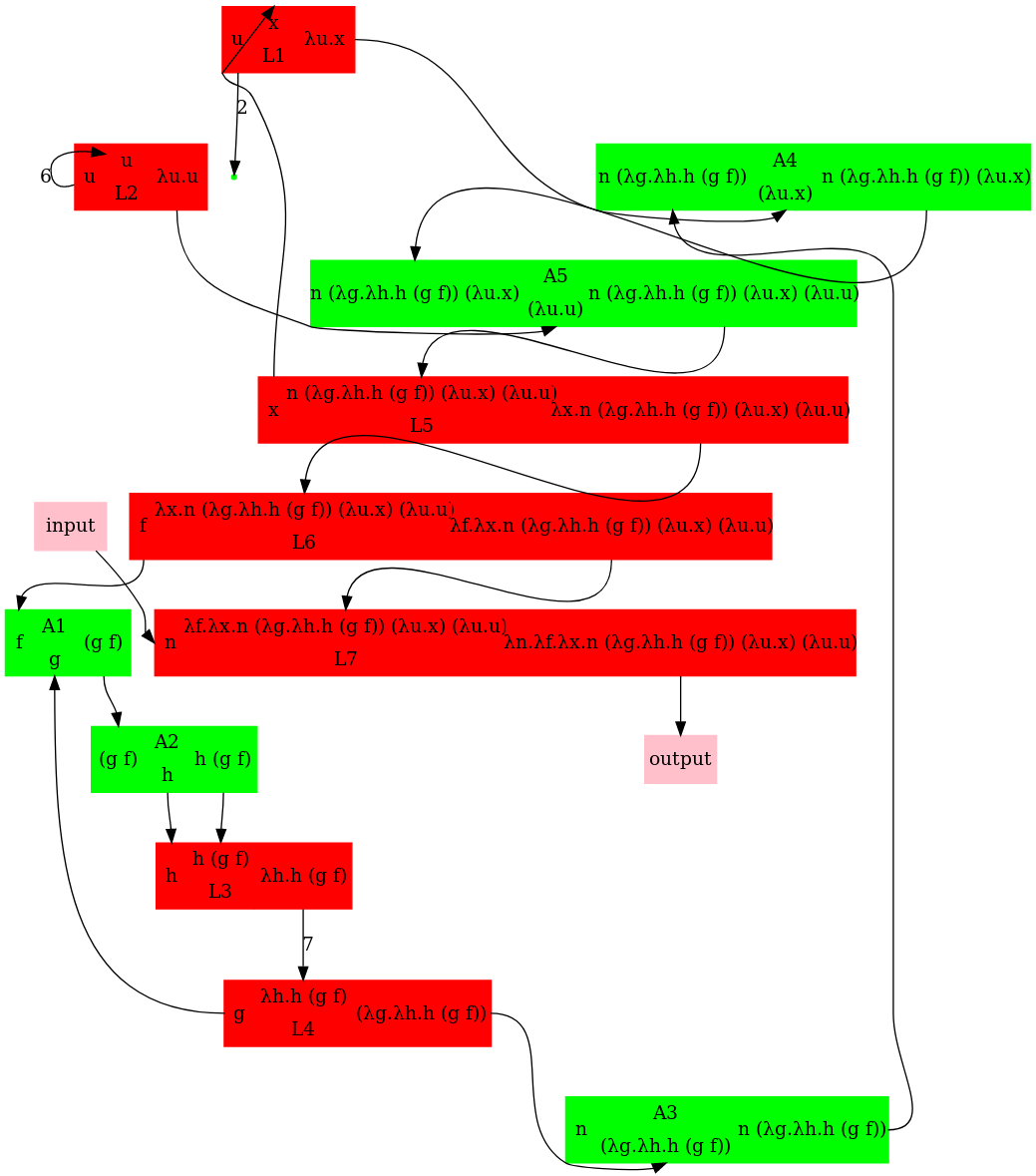
Many problems still.
(1) Arrow orientations wrong (ex. arrow from L2 to A3, corresponding to h)
(2) what is "input" and "output"?
(3) problems with port types. It's easy. The mol A a b c tells that:
Same for FI a b c.
For L a b c you have
Same for FO and FOE.
For T a and FROUT a you have
For FRIN a you have
For Arrow a b you have
Any port variable appears at most twice in a port. If it appears exactly twice, it should appear once in one of the ports li, ri, mi, and once in one of the ports lo, eo, mo. It represents an arrow directed from a port lo, mo, mo, to a port li, ri, mi. If the variable appears only once, ten it is a free arrow. So if a appears once in a li, ri, mi port, then you have to add a FRIN a, otherwise it appears in a lo, ro, mo and you have to add a FROUT a. (this explains the types of ports for FRIN, frout and T)
(4) Why A1, A2, L7, etc? A, L, FI, FO, FOE, T, Arrow, FRIN and FROUT are types of nodes. Why concatenate a node type with a numbering of nodes?
Mind that the node numbering is not part of the fatgraph. In the mol notation you may take the node numbering as the number of line, but this is only because computers are as they are. The graph rewrites don't care about the order of lines in the mol file and they should not care about the numbering of the nodes.