Created
March 24, 2017 18:38
-
-
Save AashishTiwari/cfb4082ca409d06f77201ca1dd5fdeda to your computer and use it in GitHub Desktop.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| { | |
| "cells": [ | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "# Modern NLP on OpenFDA dataset" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "#### Credits:\n", | |
| "(http://pydata.org/dc2016/schedule/presentation/11/). To view the video of the presentation on YouTube, see [here](https://www.youtube.com/watch?v=6zm9NC9uRkk)._" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "## Our Trail Map\n", | |
| "An end-to-end data science & natural language processing pipeline, starting with **raw data** and running through **preparing**, **modeling**, **visualizing**, and **analyzing** the data. We'll touch on the following points:\n", | |
| "1. A tour of the dataset\n", | |
| "1. Introduction to text processing with spaCy\n", | |
| "1. Automatic phrase modeling\n", | |
| "1. Topic modeling with LDA\n", | |
| "1. Visualizing topic models with pyLDAvis\n", | |
| "1. Word vector models with word2vec\n", | |
| "1. Visualizing word2vec with t-SNE" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "## The OpenFDA Dataset" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 1, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "import os\n", | |
| "import codecs\n", | |
| "\n", | |
| "data_directory = os.path.join('openfda_data')\n", | |
| "\n", | |
| "businesses_filepath = os.path.join(data_directory,\n", | |
| " 'drug-enforcement-0001-of-0001.json')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 2, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "2,067 Recall Reasons in the dataset.\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "import json\n", | |
| "\n", | |
| "recall_reasons = set()\n", | |
| "\n", | |
| "with open(businesses_filepath) as data_file: \n", | |
| " data = json.load(data_file)\n", | |
| " \n", | |
| " # iterate through each line (json record) in the file\n", | |
| " for business_json in data[\"results\"]: \n", | |
| " recall_reasons.add(business_json[\"reason_for_recall\"])\n", | |
| "\n", | |
| "# turn restaurant_ids into a frozenset, as we don't need to change it anymore\n", | |
| "recall_reasons_uniq = frozenset(recall_reasons)\n", | |
| "\n", | |
| "# print the number of unique restaurant ids in the dataset\n", | |
| "print(\"{:,}\".format(len(recall_reasons_uniq)), \"Recall Reasons in the dataset.\")" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Next, we will create a new file that contains only the text from recall reasons about drugs, with one reason per line in the file." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 3, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "intermediate_directory = os.path.join('d:', 'open_fda_intermediate')\n", | |
| "\n", | |
| "recall_txt_filepath = os.path.join(intermediate_directory,\n", | |
| " 'recall_reasons_all.txt')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 4, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "Text from 8,931 recall reasons in the txt file.\n", | |
| "Wall time: 280 ms\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "%%time\n", | |
| "\n", | |
| "# this is a bit time consuming - make the if statement True\n", | |
| "# if you want to execute data prep yourself.\n", | |
| "if 0 == 1:\n", | |
| " \n", | |
| " reason_count = 0\n", | |
| "\n", | |
| " # create & open a new file in write mode\n", | |
| " with codecs.open(recall_txt_filepath, 'w', encoding='utf_8') as recall_txt_file:\n", | |
| "\n", | |
| " # open the existing review json file\n", | |
| " with open(businesses_filepath) as data_file:\n", | |
| " data = json.load(data_file)\n", | |
| "\n", | |
| " for drug_details in data[\"results\"]:\n", | |
| " all_reasons = drug_details[\"reason_for_recall\"].split(';')\n", | |
| " \n", | |
| " # if this recall reason is empty or less than 5 characters dont add to final file\n", | |
| " for reason in all_reasons:\n", | |
| " if reason is None or reason is \" \" or len(reason) <= 10:\n", | |
| " continue\n", | |
| "\n", | |
| " # write the restaurant review as a line in the new file\n", | |
| " # escape newline characters in the original review text\n", | |
| " recall_txt_file.write(reason.replace('\\n', '\\\\n') + '\\n')\n", | |
| " reason_count += 1\n", | |
| "\n", | |
| " print(\"Text from {:,} recall reasons written to the new txt file.\".format(reason_count))\n", | |
| " \n", | |
| "else:\n", | |
| " \n", | |
| " with codecs.open(recall_txt_filepath, encoding='utf_8') as recall_txt_file:\n", | |
| " for reason_count, line in enumerate(recall_txt_file):\n", | |
| " pass\n", | |
| " \n", | |
| " print(\"Text from {:,} recall reasons in the txt file.\".format(reason_count + 1))" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "## spaCy — Industrial-Strength NLP in Python" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "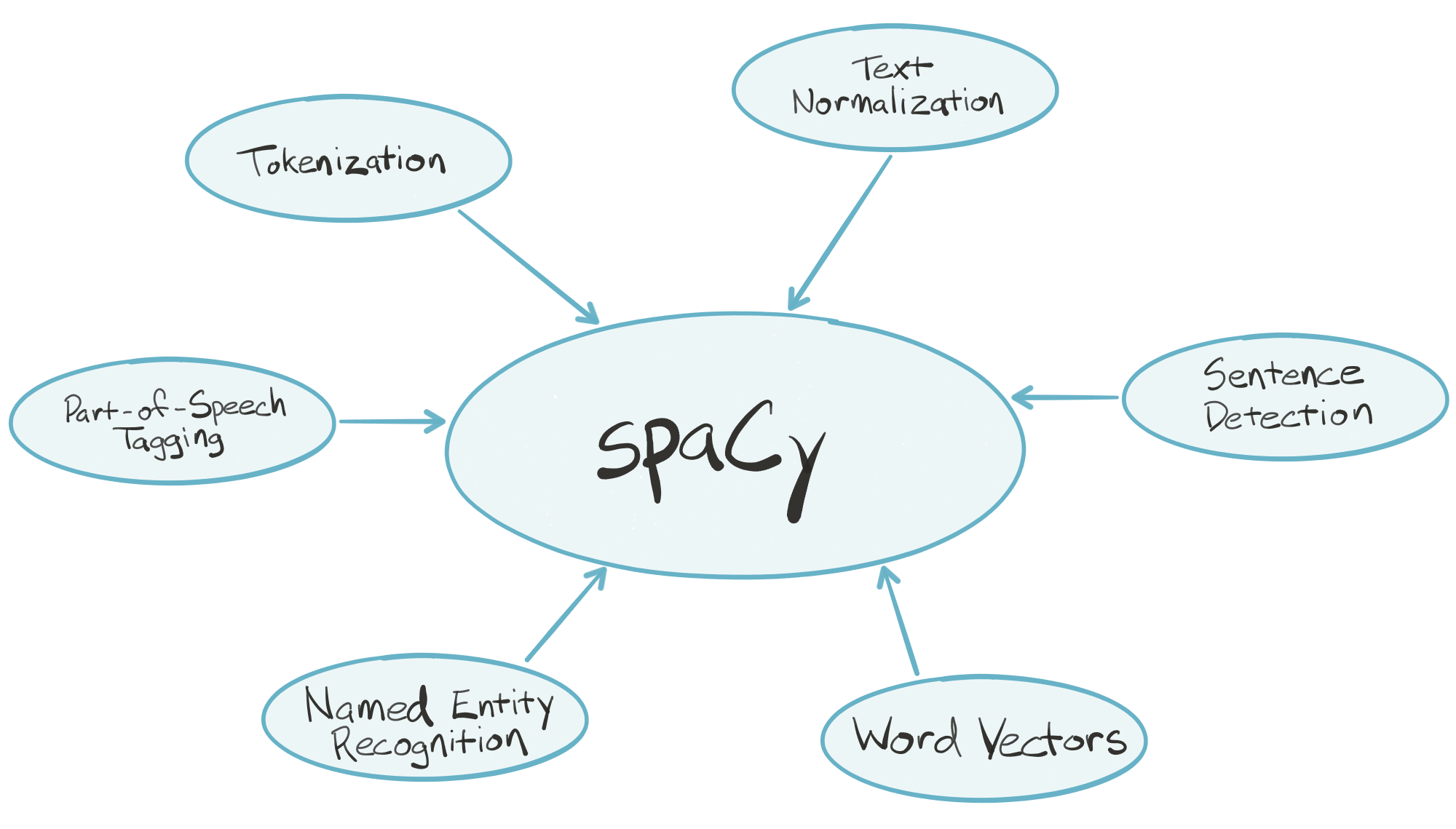" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "[**spaCy**](https://spacy.io) is an industrial-strength natural language processing (_NLP_) library for Python. spaCy's goal is to take recent advancements in natural language processing out of research papers and put them in the hands of users to build production software.\n", | |
| "\n", | |
| "spaCy handles many tasks commonly associated with building an end-to-end natural language processing pipeline:\n", | |
| "- Tokenization\n", | |
| "- Text normalization, such as lowercasing, stemming/lemmatization\n", | |
| "- Part-of-speech tagging\n", | |
| "- Syntactic dependency parsing\n", | |
| "- Sentence boundary detection\n", | |
| "- Named entity recognition and annotation\n", | |
| "\n", | |
| "In the \"batteries included\" Python tradition, spaCy contains built-in data and models which you can use out-of-the-box for processing general-purpose English language text:\n", | |
| "- Large English vocabulary, including stopword lists\n", | |
| "- Token \"probabilities\"\n", | |
| "- Word vectors\n", | |
| "\n", | |
| "spaCy is written in optimized Cython, which means it's _fast_. According to a few independent sources, it's the fastest syntactic parser available in any language. Key pieces of the spaCy parsing pipeline are written in pure C, enabling efficient multithreading (i.e., spaCy can release the _GIL_)." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 5, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "import spacy\n", | |
| "import pandas as pd\n", | |
| "import itertools as it\n", | |
| "\n", | |
| "nlp = spacy.load('en')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Let's grab a sample review to play with." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 6, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "Lack of Assurance of Sterility: Franck's Lab Inc. initiated a recall of all Sterile Human Drugs distributed between 11/21/2011 and 05/21/2012 because FDA environmental sampling revealed the presence of microorganisms and fungal growth in the clean room where sterile products were prepared. \n", | |
| "\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "with codecs.open(recall_txt_filepath, encoding='utf_8') as f:\n", | |
| " sample_reason = list(it.islice(f, 8, 9))[0]\n", | |
| " sample_reason = sample_reason.replace('\\\\n', '\\n')\n", | |
| " \n", | |
| "print(sample_reason)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Hand the review text to spaCy, and be prepared to wait..." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 7, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "Wall time: 12 ms\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "%%time\n", | |
| "parsed_reason = nlp(sample_reason)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 8, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "Lack of Assurance of Sterility: Franck's Lab Inc. initiated a recall of all Sterile Human Drugs distributed between 11/21/2011 and 05/21/2012 because FDA environmental sampling revealed the presence of microorganisms and fungal growth in the clean room where sterile products were prepared. \n", | |
| "\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "print(parsed_reason)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Looks the same! What happened under the hood?\n", | |
| "\n", | |
| "What about sentence detection and segmentation?" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 9, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "Sentence 1:\n", | |
| "Lack of Assurance of Sterility: Franck's Lab Inc. initiated a recall of all Sterile Human Drugs distributed between 11/21/2011 and 05/21/2012 because FDA environmental sampling revealed the presence of microorganisms and fungal growth in the clean room where sterile products were prepared. \n", | |
| "\n", | |
| "\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "for num, sentence in enumerate(parsed_reason.sents):\n", | |
| " print('Sentence {}:'.format(num + 1))\n", | |
| " print(sentence)\n", | |
| " print('')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "What about named entity detection?" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 10, | |
| "metadata": { | |
| "collapsed": false, | |
| "scrolled": true | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "Entity 1: FDA - ORG\n", | |
| "\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "for num, entity in enumerate(parsed_reason.ents):\n", | |
| " print('Entity {}:'.format(num + 1), entity, '-', entity.label_)\n", | |
| " print('')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "What about part of speech tagging?" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 11, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "data": { | |
| "text/html": [ | |
| "<div>\n", | |
| "<table border=\"1\" class=\"dataframe\">\n", | |
| " <thead>\n", | |
| " <tr style=\"text-align: right;\">\n", | |
| " <th></th>\n", | |
| " <th>token_text</th>\n", | |
| " <th>part_of_speech</th>\n", | |
| " </tr>\n", | |
| " </thead>\n", | |
| " <tbody>\n", | |
| " <tr>\n", | |
| " <th>0</th>\n", | |
| " <td>Lack</td>\n", | |
| " <td>NOUN</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>1</th>\n", | |
| " <td>of</td>\n", | |
| " <td>ADP</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>2</th>\n", | |
| " <td>Assurance</td>\n", | |
| " <td>PROPN</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>3</th>\n", | |
| " <td>of</td>\n", | |
| " <td>ADP</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>4</th>\n", | |
| " <td>Sterility</td>\n", | |
| " <td>NOUN</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>5</th>\n", | |
| " <td>:</td>\n", | |
| " <td>PUNCT</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>6</th>\n", | |
| " <td>Franck</td>\n", | |
| " <td>PROPN</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>7</th>\n", | |
| " <td>'s</td>\n", | |
| " <td>PART</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>8</th>\n", | |
| " <td>Lab</td>\n", | |
| " <td>PROPN</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>9</th>\n", | |
| " <td>Inc.</td>\n", | |
| " <td>PROPN</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>10</th>\n", | |
| " <td>initiated</td>\n", | |
| " <td>VERB</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>11</th>\n", | |
| " <td>a</td>\n", | |
| " <td>DET</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>12</th>\n", | |
| " <td>recall</td>\n", | |
| " <td>NOUN</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>13</th>\n", | |
| " <td>of</td>\n", | |
| " <td>ADP</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>14</th>\n", | |
| " <td>all</td>\n", | |
| " <td>DET</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>15</th>\n", | |
| " <td>Sterile</td>\n", | |
| " <td>PROPN</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>16</th>\n", | |
| " <td>Human</td>\n", | |
| " <td>PROPN</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>17</th>\n", | |
| " <td>Drugs</td>\n", | |
| " <td>PROPN</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>18</th>\n", | |
| " <td>distributed</td>\n", | |
| " <td>VERB</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>19</th>\n", | |
| " <td>between</td>\n", | |
| " <td>ADP</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>20</th>\n", | |
| " <td>11/21/2011</td>\n", | |
| " <td>NUM</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>21</th>\n", | |
| " <td>and</td>\n", | |
| " <td>CONJ</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>22</th>\n", | |
| " <td>05/21/2012</td>\n", | |
| " <td>NOUN</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>23</th>\n", | |
| " <td>because</td>\n", | |
| " <td>ADP</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>24</th>\n", | |
| " <td>FDA</td>\n", | |
| " <td>PROPN</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>25</th>\n", | |
| " <td>environmental</td>\n", | |
| " <td>ADJ</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>26</th>\n", | |
| " <td>sampling</td>\n", | |
| " <td>NOUN</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>27</th>\n", | |
| " <td>revealed</td>\n", | |
| " <td>VERB</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>28</th>\n", | |
| " <td>the</td>\n", | |
| " <td>DET</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>29</th>\n", | |
| " <td>presence</td>\n", | |
| " <td>NOUN</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>30</th>\n", | |
| " <td>of</td>\n", | |
| " <td>ADP</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>31</th>\n", | |
| " <td>microorganisms</td>\n", | |
| " <td>NOUN</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>32</th>\n", | |
| " <td>and</td>\n", | |
| " <td>CONJ</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>33</th>\n", | |
| " <td>fungal</td>\n", | |
| " <td>ADJ</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>34</th>\n", | |
| " <td>growth</td>\n", | |
| " <td>NOUN</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>35</th>\n", | |
| " <td>in</td>\n", | |
| " <td>ADP</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>36</th>\n", | |
| " <td>the</td>\n", | |
| " <td>DET</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>37</th>\n", | |
| " <td>clean</td>\n", | |
| " <td>ADJ</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>38</th>\n", | |
| " <td>room</td>\n", | |
| " <td>NOUN</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>39</th>\n", | |
| " <td>where</td>\n", | |
| " <td>ADV</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>40</th>\n", | |
| " <td>sterile</td>\n", | |
| " <td>ADJ</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>41</th>\n", | |
| " <td>products</td>\n", | |
| " <td>NOUN</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>42</th>\n", | |
| " <td>were</td>\n", | |
| " <td>VERB</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>43</th>\n", | |
| " <td>prepared</td>\n", | |
| " <td>ADJ</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>44</th>\n", | |
| " <td>.</td>\n", | |
| " <td>PUNCT</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>45</th>\n", | |
| " <td>\\n</td>\n", | |
| " <td>SPACE</td>\n", | |
| " </tr>\n", | |
| " </tbody>\n", | |
| "</table>\n", | |
| "</div>" | |
| ], | |
| "text/plain": [ | |
| " token_text part_of_speech\n", | |
| "0 Lack NOUN\n", | |
| "1 of ADP\n", | |
| "2 Assurance PROPN\n", | |
| "3 of ADP\n", | |
| "4 Sterility NOUN\n", | |
| "5 : PUNCT\n", | |
| "6 Franck PROPN\n", | |
| "7 's PART\n", | |
| "8 Lab PROPN\n", | |
| "9 Inc. PROPN\n", | |
| "10 initiated VERB\n", | |
| "11 a DET\n", | |
| "12 recall NOUN\n", | |
| "13 of ADP\n", | |
| "14 all DET\n", | |
| "15 Sterile PROPN\n", | |
| "16 Human PROPN\n", | |
| "17 Drugs PROPN\n", | |
| "18 distributed VERB\n", | |
| "19 between ADP\n", | |
| "20 11/21/2011 NUM\n", | |
| "21 and CONJ\n", | |
| "22 05/21/2012 NOUN\n", | |
| "23 because ADP\n", | |
| "24 FDA PROPN\n", | |
| "25 environmental ADJ\n", | |
| "26 sampling NOUN\n", | |
| "27 revealed VERB\n", | |
| "28 the DET\n", | |
| "29 presence NOUN\n", | |
| "30 of ADP\n", | |
| "31 microorganisms NOUN\n", | |
| "32 and CONJ\n", | |
| "33 fungal ADJ\n", | |
| "34 growth NOUN\n", | |
| "35 in ADP\n", | |
| "36 the DET\n", | |
| "37 clean ADJ\n", | |
| "38 room NOUN\n", | |
| "39 where ADV\n", | |
| "40 sterile ADJ\n", | |
| "41 products NOUN\n", | |
| "42 were VERB\n", | |
| "43 prepared ADJ\n", | |
| "44 . PUNCT\n", | |
| "45 \\n SPACE" | |
| ] | |
| }, | |
| "execution_count": 11, | |
| "metadata": {}, | |
| "output_type": "execute_result" | |
| } | |
| ], | |
| "source": [ | |
| "token_text = [token.orth_ for token in parsed_reason]\n", | |
| "token_pos = [token.pos_ for token in parsed_reason]\n", | |
| "\n", | |
| "pd.DataFrame(list(zip(token_text, token_pos)),\n", | |
| " columns=['token_text', 'part_of_speech'])" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "What about text normalization, like stemming/lemmatization and shape analysis?" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 12, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "data": { | |
| "text/html": [ | |
| "<div>\n", | |
| "<table border=\"1\" class=\"dataframe\">\n", | |
| " <thead>\n", | |
| " <tr style=\"text-align: right;\">\n", | |
| " <th></th>\n", | |
| " <th>token_text</th>\n", | |
| " <th>token_lemma</th>\n", | |
| " <th>token_shape</th>\n", | |
| " </tr>\n", | |
| " </thead>\n", | |
| " <tbody>\n", | |
| " <tr>\n", | |
| " <th>0</th>\n", | |
| " <td>Lack</td>\n", | |
| " <td>lack</td>\n", | |
| " <td>Xxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>1</th>\n", | |
| " <td>of</td>\n", | |
| " <td>of</td>\n", | |
| " <td>xx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>2</th>\n", | |
| " <td>Assurance</td>\n", | |
| " <td>assurance</td>\n", | |
| " <td>Xxxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>3</th>\n", | |
| " <td>of</td>\n", | |
| " <td>of</td>\n", | |
| " <td>xx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>4</th>\n", | |
| " <td>Sterility</td>\n", | |
| " <td>sterility</td>\n", | |
| " <td>Xxxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>5</th>\n", | |
| " <td>:</td>\n", | |
| " <td>:</td>\n", | |
| " <td>:</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>6</th>\n", | |
| " <td>Franck</td>\n", | |
| " <td>franck</td>\n", | |
| " <td>Xxxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>7</th>\n", | |
| " <td>'s</td>\n", | |
| " <td>'s</td>\n", | |
| " <td>'x</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>8</th>\n", | |
| " <td>Lab</td>\n", | |
| " <td>lab</td>\n", | |
| " <td>Xxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>9</th>\n", | |
| " <td>Inc.</td>\n", | |
| " <td>inc.</td>\n", | |
| " <td>Xxx.</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>10</th>\n", | |
| " <td>initiated</td>\n", | |
| " <td>initiate</td>\n", | |
| " <td>xxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>11</th>\n", | |
| " <td>a</td>\n", | |
| " <td>a</td>\n", | |
| " <td>x</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>12</th>\n", | |
| " <td>recall</td>\n", | |
| " <td>recall</td>\n", | |
| " <td>xxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>13</th>\n", | |
| " <td>of</td>\n", | |
| " <td>of</td>\n", | |
| " <td>xx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>14</th>\n", | |
| " <td>all</td>\n", | |
| " <td>all</td>\n", | |
| " <td>xxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>15</th>\n", | |
| " <td>Sterile</td>\n", | |
| " <td>sterile</td>\n", | |
| " <td>Xxxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>16</th>\n", | |
| " <td>Human</td>\n", | |
| " <td>human</td>\n", | |
| " <td>Xxxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>17</th>\n", | |
| " <td>Drugs</td>\n", | |
| " <td>drugs</td>\n", | |
| " <td>Xxxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>18</th>\n", | |
| " <td>distributed</td>\n", | |
| " <td>distribute</td>\n", | |
| " <td>xxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>19</th>\n", | |
| " <td>between</td>\n", | |
| " <td>between</td>\n", | |
| " <td>xxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>20</th>\n", | |
| " <td>11/21/2011</td>\n", | |
| " <td>11/21/2011</td>\n", | |
| " <td>dd/dd/dddd</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>21</th>\n", | |
| " <td>and</td>\n", | |
| " <td>and</td>\n", | |
| " <td>xxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>22</th>\n", | |
| " <td>05/21/2012</td>\n", | |
| " <td>05/21/2012</td>\n", | |
| " <td>dd/dd/dddd</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>23</th>\n", | |
| " <td>because</td>\n", | |
| " <td>because</td>\n", | |
| " <td>xxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>24</th>\n", | |
| " <td>FDA</td>\n", | |
| " <td>fda</td>\n", | |
| " <td>XXX</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>25</th>\n", | |
| " <td>environmental</td>\n", | |
| " <td>environmental</td>\n", | |
| " <td>xxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>26</th>\n", | |
| " <td>sampling</td>\n", | |
| " <td>sampling</td>\n", | |
| " <td>xxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>27</th>\n", | |
| " <td>revealed</td>\n", | |
| " <td>reveal</td>\n", | |
| " <td>xxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>28</th>\n", | |
| " <td>the</td>\n", | |
| " <td>the</td>\n", | |
| " <td>xxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>29</th>\n", | |
| " <td>presence</td>\n", | |
| " <td>presence</td>\n", | |
| " <td>xxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>30</th>\n", | |
| " <td>of</td>\n", | |
| " <td>of</td>\n", | |
| " <td>xx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>31</th>\n", | |
| " <td>microorganisms</td>\n", | |
| " <td>microorganism</td>\n", | |
| " <td>xxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>32</th>\n", | |
| " <td>and</td>\n", | |
| " <td>and</td>\n", | |
| " <td>xxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>33</th>\n", | |
| " <td>fungal</td>\n", | |
| " <td>fungal</td>\n", | |
| " <td>xxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>34</th>\n", | |
| " <td>growth</td>\n", | |
| " <td>growth</td>\n", | |
| " <td>xxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>35</th>\n", | |
| " <td>in</td>\n", | |
| " <td>in</td>\n", | |
| " <td>xx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>36</th>\n", | |
| " <td>the</td>\n", | |
| " <td>the</td>\n", | |
| " <td>xxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>37</th>\n", | |
| " <td>clean</td>\n", | |
| " <td>clean</td>\n", | |
| " <td>xxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>38</th>\n", | |
| " <td>room</td>\n", | |
| " <td>room</td>\n", | |
| " <td>xxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>39</th>\n", | |
| " <td>where</td>\n", | |
| " <td>where</td>\n", | |
| " <td>xxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>40</th>\n", | |
| " <td>sterile</td>\n", | |
| " <td>sterile</td>\n", | |
| " <td>xxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>41</th>\n", | |
| " <td>products</td>\n", | |
| " <td>product</td>\n", | |
| " <td>xxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>42</th>\n", | |
| " <td>were</td>\n", | |
| " <td>be</td>\n", | |
| " <td>xxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>43</th>\n", | |
| " <td>prepared</td>\n", | |
| " <td>prepared</td>\n", | |
| " <td>xxxx</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>44</th>\n", | |
| " <td>.</td>\n", | |
| " <td>.</td>\n", | |
| " <td>.</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>45</th>\n", | |
| " <td>\\n</td>\n", | |
| " <td>\\n</td>\n", | |
| " <td>\\n</td>\n", | |
| " </tr>\n", | |
| " </tbody>\n", | |
| "</table>\n", | |
| "</div>" | |
| ], | |
| "text/plain": [ | |
| " token_text token_lemma token_shape\n", | |
| "0 Lack lack Xxxx\n", | |
| "1 of of xx\n", | |
| "2 Assurance assurance Xxxxx\n", | |
| "3 of of xx\n", | |
| "4 Sterility sterility Xxxxx\n", | |
| "5 : : :\n", | |
| "6 Franck franck Xxxxx\n", | |
| "7 's 's 'x\n", | |
| "8 Lab lab Xxx\n", | |
| "9 Inc. inc. Xxx.\n", | |
| "10 initiated initiate xxxx\n", | |
| "11 a a x\n", | |
| "12 recall recall xxxx\n", | |
| "13 of of xx\n", | |
| "14 all all xxx\n", | |
| "15 Sterile sterile Xxxxx\n", | |
| "16 Human human Xxxxx\n", | |
| "17 Drugs drugs Xxxxx\n", | |
| "18 distributed distribute xxxx\n", | |
| "19 between between xxxx\n", | |
| "20 11/21/2011 11/21/2011 dd/dd/dddd\n", | |
| "21 and and xxx\n", | |
| "22 05/21/2012 05/21/2012 dd/dd/dddd\n", | |
| "23 because because xxxx\n", | |
| "24 FDA fda XXX\n", | |
| "25 environmental environmental xxxx\n", | |
| "26 sampling sampling xxxx\n", | |
| "27 revealed reveal xxxx\n", | |
| "28 the the xxx\n", | |
| "29 presence presence xxxx\n", | |
| "30 of of xx\n", | |
| "31 microorganisms microorganism xxxx\n", | |
| "32 and and xxx\n", | |
| "33 fungal fungal xxxx\n", | |
| "34 growth growth xxxx\n", | |
| "35 in in xx\n", | |
| "36 the the xxx\n", | |
| "37 clean clean xxxx\n", | |
| "38 room room xxxx\n", | |
| "39 where where xxxx\n", | |
| "40 sterile sterile xxxx\n", | |
| "41 products product xxxx\n", | |
| "42 were be xxxx\n", | |
| "43 prepared prepared xxxx\n", | |
| "44 . . .\n", | |
| "45 \\n \\n \\n" | |
| ] | |
| }, | |
| "execution_count": 12, | |
| "metadata": {}, | |
| "output_type": "execute_result" | |
| } | |
| ], | |
| "source": [ | |
| "token_lemma = [token.lemma_ for token in parsed_reason]\n", | |
| "token_shape = [token.shape_ for token in parsed_reason]\n", | |
| "\n", | |
| "pd.DataFrame(list(zip(token_text, token_lemma, token_shape)),\n", | |
| " columns=['token_text', 'token_lemma', 'token_shape'])" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "What about token-level entity analysis?" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 13, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "data": { | |
| "text/html": [ | |
| "<div>\n", | |
| "<table border=\"1\" class=\"dataframe\">\n", | |
| " <thead>\n", | |
| " <tr style=\"text-align: right;\">\n", | |
| " <th></th>\n", | |
| " <th>token_text</th>\n", | |
| " <th>entity_type</th>\n", | |
| " <th>inside_outside_begin</th>\n", | |
| " </tr>\n", | |
| " </thead>\n", | |
| " <tbody>\n", | |
| " <tr>\n", | |
| " <th>0</th>\n", | |
| " <td>Lack</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>1</th>\n", | |
| " <td>of</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>2</th>\n", | |
| " <td>Assurance</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>3</th>\n", | |
| " <td>of</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>4</th>\n", | |
| " <td>Sterility</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>5</th>\n", | |
| " <td>:</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>6</th>\n", | |
| " <td>Franck</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>7</th>\n", | |
| " <td>'s</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>8</th>\n", | |
| " <td>Lab</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>9</th>\n", | |
| " <td>Inc.</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>10</th>\n", | |
| " <td>initiated</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>11</th>\n", | |
| " <td>a</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>12</th>\n", | |
| " <td>recall</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>13</th>\n", | |
| " <td>of</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>14</th>\n", | |
| " <td>all</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>15</th>\n", | |
| " <td>Sterile</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>16</th>\n", | |
| " <td>Human</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>17</th>\n", | |
| " <td>Drugs</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>18</th>\n", | |
| " <td>distributed</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>19</th>\n", | |
| " <td>between</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>20</th>\n", | |
| " <td>11/21/2011</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>21</th>\n", | |
| " <td>and</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>22</th>\n", | |
| " <td>05/21/2012</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>23</th>\n", | |
| " <td>because</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>24</th>\n", | |
| " <td>FDA</td>\n", | |
| " <td>ORG</td>\n", | |
| " <td>B</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>25</th>\n", | |
| " <td>environmental</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>26</th>\n", | |
| " <td>sampling</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>27</th>\n", | |
| " <td>revealed</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>28</th>\n", | |
| " <td>the</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>29</th>\n", | |
| " <td>presence</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>30</th>\n", | |
| " <td>of</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>31</th>\n", | |
| " <td>microorganisms</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>32</th>\n", | |
| " <td>and</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>33</th>\n", | |
| " <td>fungal</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>34</th>\n", | |
| " <td>growth</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>35</th>\n", | |
| " <td>in</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>36</th>\n", | |
| " <td>the</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>37</th>\n", | |
| " <td>clean</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>38</th>\n", | |
| " <td>room</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>39</th>\n", | |
| " <td>where</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>40</th>\n", | |
| " <td>sterile</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>41</th>\n", | |
| " <td>products</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>42</th>\n", | |
| " <td>were</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>43</th>\n", | |
| " <td>prepared</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>44</th>\n", | |
| " <td>.</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>45</th>\n", | |
| " <td>\\n</td>\n", | |
| " <td></td>\n", | |
| " <td>O</td>\n", | |
| " </tr>\n", | |
| " </tbody>\n", | |
| "</table>\n", | |
| "</div>" | |
| ], | |
| "text/plain": [ | |
| " token_text entity_type inside_outside_begin\n", | |
| "0 Lack O\n", | |
| "1 of O\n", | |
| "2 Assurance O\n", | |
| "3 of O\n", | |
| "4 Sterility O\n", | |
| "5 : O\n", | |
| "6 Franck O\n", | |
| "7 's O\n", | |
| "8 Lab O\n", | |
| "9 Inc. O\n", | |
| "10 initiated O\n", | |
| "11 a O\n", | |
| "12 recall O\n", | |
| "13 of O\n", | |
| "14 all O\n", | |
| "15 Sterile O\n", | |
| "16 Human O\n", | |
| "17 Drugs O\n", | |
| "18 distributed O\n", | |
| "19 between O\n", | |
| "20 11/21/2011 O\n", | |
| "21 and O\n", | |
| "22 05/21/2012 O\n", | |
| "23 because O\n", | |
| "24 FDA ORG B\n", | |
| "25 environmental O\n", | |
| "26 sampling O\n", | |
| "27 revealed O\n", | |
| "28 the O\n", | |
| "29 presence O\n", | |
| "30 of O\n", | |
| "31 microorganisms O\n", | |
| "32 and O\n", | |
| "33 fungal O\n", | |
| "34 growth O\n", | |
| "35 in O\n", | |
| "36 the O\n", | |
| "37 clean O\n", | |
| "38 room O\n", | |
| "39 where O\n", | |
| "40 sterile O\n", | |
| "41 products O\n", | |
| "42 were O\n", | |
| "43 prepared O\n", | |
| "44 . O\n", | |
| "45 \\n O" | |
| ] | |
| }, | |
| "execution_count": 13, | |
| "metadata": {}, | |
| "output_type": "execute_result" | |
| } | |
| ], | |
| "source": [ | |
| "token_entity_type = [token.ent_type_ for token in parsed_reason]\n", | |
| "token_entity_iob = [token.ent_iob_ for token in parsed_reason]\n", | |
| "\n", | |
| "pd.DataFrame(list(zip(token_text, token_entity_type, token_entity_iob)),\n", | |
| " columns=['token_text', 'entity_type', 'inside_outside_begin'])" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "What about a variety of other token-level attributes, such as the relative frequency of tokens, and whether or not a token matches any of these categories?\n", | |
| "- stopword\n", | |
| "- punctuation\n", | |
| "- whitespace\n", | |
| "- represents a number\n", | |
| "- whether or not the token is included in spaCy's default vocabulary?" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 15, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "data": { | |
| "text/html": [ | |
| "<div>\n", | |
| "<table border=\"1\" class=\"dataframe\">\n", | |
| " <thead>\n", | |
| " <tr style=\"text-align: right;\">\n", | |
| " <th></th>\n", | |
| " <th>text</th>\n", | |
| " <th>log_probability</th>\n", | |
| " <th>stop?</th>\n", | |
| " <th>punctuation?</th>\n", | |
| " <th>whitespace?</th>\n", | |
| " <th>number?</th>\n", | |
| " <th>out of vocab.?</th>\n", | |
| " </tr>\n", | |
| " </thead>\n", | |
| " <tbody>\n", | |
| " <tr>\n", | |
| " <th>0</th>\n", | |
| " <td>Lack</td>\n", | |
| " <td>-12.744525</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>1</th>\n", | |
| " <td>of</td>\n", | |
| " <td>-4.275874</td>\n", | |
| " <td>Yes</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>2</th>\n", | |
| " <td>Assurance</td>\n", | |
| " <td>-15.678530</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>3</th>\n", | |
| " <td>of</td>\n", | |
| " <td>-4.275874</td>\n", | |
| " <td>Yes</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>4</th>\n", | |
| " <td>Sterility</td>\n", | |
| " <td>-18.106260</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>5</th>\n", | |
| " <td>:</td>\n", | |
| " <td>-6.128876</td>\n", | |
| " <td></td>\n", | |
| " <td>Yes</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>6</th>\n", | |
| " <td>Franck</td>\n", | |
| " <td>-16.658981</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>7</th>\n", | |
| " <td>'s</td>\n", | |
| " <td>-4.830559</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>8</th>\n", | |
| " <td>Lab</td>\n", | |
| " <td>-13.243361</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>9</th>\n", | |
| " <td>Inc.</td>\n", | |
| " <td>-13.148737</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>10</th>\n", | |
| " <td>initiated</td>\n", | |
| " <td>-12.858611</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>11</th>\n", | |
| " <td>a</td>\n", | |
| " <td>-3.929788</td>\n", | |
| " <td>Yes</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>12</th>\n", | |
| " <td>recall</td>\n", | |
| " <td>-10.451101</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>13</th>\n", | |
| " <td>of</td>\n", | |
| " <td>-4.275874</td>\n", | |
| " <td>Yes</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>14</th>\n", | |
| " <td>all</td>\n", | |
| " <td>-5.936641</td>\n", | |
| " <td>Yes</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>15</th>\n", | |
| " <td>Sterile</td>\n", | |
| " <td>-16.518122</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>16</th>\n", | |
| " <td>Human</td>\n", | |
| " <td>-11.457912</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>17</th>\n", | |
| " <td>Drugs</td>\n", | |
| " <td>-12.628650</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>18</th>\n", | |
| " <td>distributed</td>\n", | |
| " <td>-12.064220</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>19</th>\n", | |
| " <td>between</td>\n", | |
| " <td>-8.106386</td>\n", | |
| " <td>Yes</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>20</th>\n", | |
| " <td>11/21/2011</td>\n", | |
| " <td>-19.502029</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td>Yes</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>21</th>\n", | |
| " <td>and</td>\n", | |
| " <td>-4.113108</td>\n", | |
| " <td>Yes</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>22</th>\n", | |
| " <td>05/21/2012</td>\n", | |
| " <td>-19.502029</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td>Yes</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>23</th>\n", | |
| " <td>because</td>\n", | |
| " <td>-6.349620</td>\n", | |
| " <td>Yes</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>24</th>\n", | |
| " <td>FDA</td>\n", | |
| " <td>-12.629486</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>25</th>\n", | |
| " <td>environmental</td>\n", | |
| " <td>-11.756105</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>26</th>\n", | |
| " <td>sampling</td>\n", | |
| " <td>-12.960810</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>27</th>\n", | |
| " <td>revealed</td>\n", | |
| " <td>-11.573030</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>28</th>\n", | |
| " <td>the</td>\n", | |
| " <td>-3.528767</td>\n", | |
| " <td>Yes</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>29</th>\n", | |
| " <td>presence</td>\n", | |
| " <td>-10.912554</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>30</th>\n", | |
| " <td>of</td>\n", | |
| " <td>-4.275874</td>\n", | |
| " <td>Yes</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>31</th>\n", | |
| " <td>microorganisms</td>\n", | |
| " <td>-15.163286</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>32</th>\n", | |
| " <td>and</td>\n", | |
| " <td>-4.113108</td>\n", | |
| " <td>Yes</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>33</th>\n", | |
| " <td>fungal</td>\n", | |
| " <td>-13.978355</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>34</th>\n", | |
| " <td>growth</td>\n", | |
| " <td>-10.614445</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>35</th>\n", | |
| " <td>in</td>\n", | |
| " <td>-4.619072</td>\n", | |
| " <td>Yes</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>36</th>\n", | |
| " <td>the</td>\n", | |
| " <td>-3.528767</td>\n", | |
| " <td>Yes</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>37</th>\n", | |
| " <td>clean</td>\n", | |
| " <td>-9.471311</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>38</th>\n", | |
| " <td>room</td>\n", | |
| " <td>-8.814630</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>39</th>\n", | |
| " <td>where</td>\n", | |
| " <td>-7.146170</td>\n", | |
| " <td>Yes</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>40</th>\n", | |
| " <td>sterile</td>\n", | |
| " <td>-12.929901</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>41</th>\n", | |
| " <td>products</td>\n", | |
| " <td>-10.061659</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>42</th>\n", | |
| " <td>were</td>\n", | |
| " <td>-6.673175</td>\n", | |
| " <td>Yes</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>43</th>\n", | |
| " <td>prepared</td>\n", | |
| " <td>-10.667645</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>44</th>\n", | |
| " <td>.</td>\n", | |
| " <td>-3.067898</td>\n", | |
| " <td></td>\n", | |
| " <td>Yes</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>45</th>\n", | |
| " <td>\\n</td>\n", | |
| " <td>-6.050651</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " <td>Yes</td>\n", | |
| " <td></td>\n", | |
| " <td></td>\n", | |
| " </tr>\n", | |
| " </tbody>\n", | |
| "</table>\n", | |
| "</div>" | |
| ], | |
| "text/plain": [ | |
| " text log_probability stop? punctuation? whitespace? number? \\\n", | |
| "0 Lack -12.744525 \n", | |
| "1 of -4.275874 Yes \n", | |
| "2 Assurance -15.678530 \n", | |
| "3 of -4.275874 Yes \n", | |
| "4 Sterility -18.106260 \n", | |
| "5 : -6.128876 Yes \n", | |
| "6 Franck -16.658981 \n", | |
| "7 's -4.830559 \n", | |
| "8 Lab -13.243361 \n", | |
| "9 Inc. -13.148737 \n", | |
| "10 initiated -12.858611 \n", | |
| "11 a -3.929788 Yes \n", | |
| "12 recall -10.451101 \n", | |
| "13 of -4.275874 Yes \n", | |
| "14 all -5.936641 Yes \n", | |
| "15 Sterile -16.518122 \n", | |
| "16 Human -11.457912 \n", | |
| "17 Drugs -12.628650 \n", | |
| "18 distributed -12.064220 \n", | |
| "19 between -8.106386 Yes \n", | |
| "20 11/21/2011 -19.502029 \n", | |
| "21 and -4.113108 Yes \n", | |
| "22 05/21/2012 -19.502029 \n", | |
| "23 because -6.349620 Yes \n", | |
| "24 FDA -12.629486 \n", | |
| "25 environmental -11.756105 \n", | |
| "26 sampling -12.960810 \n", | |
| "27 revealed -11.573030 \n", | |
| "28 the -3.528767 Yes \n", | |
| "29 presence -10.912554 \n", | |
| "30 of -4.275874 Yes \n", | |
| "31 microorganisms -15.163286 \n", | |
| "32 and -4.113108 Yes \n", | |
| "33 fungal -13.978355 \n", | |
| "34 growth -10.614445 \n", | |
| "35 in -4.619072 Yes \n", | |
| "36 the -3.528767 Yes \n", | |
| "37 clean -9.471311 \n", | |
| "38 room -8.814630 \n", | |
| "39 where -7.146170 Yes \n", | |
| "40 sterile -12.929901 \n", | |
| "41 products -10.061659 \n", | |
| "42 were -6.673175 Yes \n", | |
| "43 prepared -10.667645 \n", | |
| "44 . -3.067898 Yes \n", | |
| "45 \\n -6.050651 Yes \n", | |
| "\n", | |
| " out of vocab.? \n", | |
| "0 \n", | |
| "1 \n", | |
| "2 \n", | |
| "3 \n", | |
| "4 \n", | |
| "5 \n", | |
| "6 \n", | |
| "7 \n", | |
| "8 \n", | |
| "9 \n", | |
| "10 \n", | |
| "11 \n", | |
| "12 \n", | |
| "13 \n", | |
| "14 \n", | |
| "15 \n", | |
| "16 \n", | |
| "17 \n", | |
| "18 \n", | |
| "19 \n", | |
| "20 Yes \n", | |
| "21 \n", | |
| "22 Yes \n", | |
| "23 \n", | |
| "24 \n", | |
| "25 \n", | |
| "26 \n", | |
| "27 \n", | |
| "28 \n", | |
| "29 \n", | |
| "30 \n", | |
| "31 \n", | |
| "32 \n", | |
| "33 \n", | |
| "34 \n", | |
| "35 \n", | |
| "36 \n", | |
| "37 \n", | |
| "38 \n", | |
| "39 \n", | |
| "40 \n", | |
| "41 \n", | |
| "42 \n", | |
| "43 \n", | |
| "44 \n", | |
| "45 " | |
| ] | |
| }, | |
| "execution_count": 15, | |
| "metadata": {}, | |
| "output_type": "execute_result" | |
| } | |
| ], | |
| "source": [ | |
| "token_attributes = [(token.orth_,\n", | |
| " token.prob,\n", | |
| " token.is_stop,\n", | |
| " token.is_punct,\n", | |
| " token.is_space,\n", | |
| " token.like_num,\n", | |
| " token.is_oov)\n", | |
| " for token in parsed_reason]\n", | |
| "\n", | |
| "df = pd.DataFrame(token_attributes,\n", | |
| " columns=['text',\n", | |
| " 'log_probability',\n", | |
| " 'stop?',\n", | |
| " 'punctuation?',\n", | |
| " 'whitespace?',\n", | |
| " 'number?',\n", | |
| " 'out of vocab.?'])\n", | |
| "\n", | |
| "df.loc[:, 'stop?':'out of vocab.?'] = (df.loc[:, 'stop?':'out of vocab.?']\n", | |
| " .applymap(lambda x: 'Yes' if x else ''))\n", | |
| " \n", | |
| "df" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "If the text you'd like to process is general-purpose English language text (i.e., not domain-specific, like medical literature), spaCy is ready to use out-of-the-box.\n", | |
| "\n", | |
| "I think it will eventually become a core part of the Python data science ecosystem — it will do for natural language computing what other great libraries have done for numerical computing." | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "## Phrase Modeling" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "_Phrase modeling_ is another approach to learning combinations of tokens that together represent meaningful multi-word concepts. We can develop phrase models by looping over the the words in our reviews and looking for words that _co-occur_ (i.e., appear one after another) together much more frequently than you would expect them to by random chance. The formula our phrase models will use to determine whether two tokens $A$ and $B$ constitute a phrase is:\n", | |
| "\n", | |
| "$$\\frac{count(A\\ B) - count_{min}}{count(A) * count(B)} * N > threshold$$\n", | |
| "\n", | |
| "...where:\n", | |
| "* $count(A)$ is the number of times token $A$ appears in the corpus\n", | |
| "* $count(B)$ is the number of times token $B$ appears in the corpus\n", | |
| "* $count(A\\ B)$ is the number of times the tokens $A\\ B$ appear in the corpus *in order*\n", | |
| "* $N$ is the total size of the corpus vocabulary\n", | |
| "* $count_{min}$ is a user-defined parameter to ensure that accepted phrases occur a minimum number of times\n", | |
| "* $threshold$ is a user-defined parameter to control how strong of a relationship between two tokens the model requires before accepting them as a phrase\n", | |
| "\n", | |
| "Once our phrase model has been trained on our corpus, we can apply it to new text. When our model encounters two tokens in new text that identifies as a phrase, it will merge the two into a single new token.\n", | |
| "\n", | |
| "Phrase modeling is superficially similar to named entity detection in that you would expect named entities to become phrases in the model (so _new york_ would become *new\\_york*). But you would also expect multi-word expressions that represent common concepts, but aren't specifically named entities (such as _happy hour_) to also become phrases in the model.\n", | |
| "\n", | |
| "We turn to the indispensible [**gensim**](https://radimrehurek.com/gensim/index.html) library to help us with phrase modeling — the [**Phrases**](https://radimrehurek.com/gensim/models/phrases.html) class in particular." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 16, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stderr", | |
| "output_type": "stream", | |
| "text": [ | |
| "C:\\Users\\aashis_tiwari\\AppData\\Local\\Continuum\\Anaconda3\\envs\\tensorflow\\lib\\site-packages\\gensim\\utils.py:855: UserWarning: detected Windows; aliasing chunkize to chunkize_serial\n", | |
| " warnings.warn(\"detected Windows; aliasing chunkize to chunkize_serial\")\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "from gensim.models import Phrases\n", | |
| "from gensim.models.word2vec import LineSentence" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "As we're performing phrase modeling, we'll be doing some iterative data transformation at the same time. Our roadmap for data preparation includes:\n", | |
| "\n", | |
| "1. Segment text of complete reviews into sentences & normalize text\n", | |
| "1. First-order phrase modeling $\\rightarrow$ _apply first-order phrase model to transform sentences_\n", | |
| "1. Second-order phrase modeling $\\rightarrow$ _apply second-order phrase model to transform sentences_\n", | |
| "1. Apply text normalization and second-order phrase model to text of complete reviews\n", | |
| "\n", | |
| "We'll use this transformed data as the input for some higher-level modeling approaches in the following sections." | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "First, let's define a few helper functions that we'll use for text normalization. In particular, the `lemmatized_sentence_corpus` generator function will use spaCy to:\n", | |
| "- Iterate over the 1M reviews in the `review_txt_all.txt` we created before\n", | |
| "- Segment the reviews into individual sentences\n", | |
| "- Remove punctuation and excess whitespace\n", | |
| "- Lemmatize the text\n", | |
| "\n", | |
| "... and do so efficiently in parallel, thanks to spaCy's `nlp.pipe()` function." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 17, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "def punct_space(token):\n", | |
| " \"\"\"\n", | |
| " helper function to eliminate tokens\n", | |
| " that are pure punctuation or whitespace\n", | |
| " \"\"\"\n", | |
| " \n", | |
| " return token.is_punct or token.is_space\n", | |
| "\n", | |
| "def line_review(filename):\n", | |
| " \"\"\"\n", | |
| " generator function to read in reviews from the file\n", | |
| " and un-escape the original line breaks in the text\n", | |
| " \"\"\"\n", | |
| " \n", | |
| " with codecs.open(filename, encoding='utf_8') as f:\n", | |
| " for review in f:\n", | |
| " yield review.replace('\\\\n', '\\n')\n", | |
| " \n", | |
| "def lemmatized_sentence_corpus(filename):\n", | |
| " \"\"\"\n", | |
| " generator function to use spaCy to parse reviews,\n", | |
| " lemmatize the text, and yield sentences\n", | |
| " \"\"\"\n", | |
| " \n", | |
| " for parsed_reason in nlp.pipe(line_review(filename),\n", | |
| " batch_size=10000, n_threads=4):\n", | |
| " \n", | |
| " for sent in parsed_reason.sents:\n", | |
| " yield u' '.join([token.lemma_ for token in sent\n", | |
| " if not punct_space(token)])" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 18, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "unigram_sentences_filepath = os.path.join(intermediate_directory,\n", | |
| " 'unigram_sentences_all.txt')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Let's use the `lemmatized_sentence_corpus` generator to loop over the original review text, segmenting the reviews into individual sentences and normalizing the text. We'll write this data back out to a new file (`unigram_sentences_all`), with one normalized sentence per line. We'll use this data for learning our phrase models." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 19, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "Wall time: 0 ns\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "%%time\n", | |
| "\n", | |
| "# this is a bit time consuming - make the if statement True\n", | |
| "# if you want to execute data prep yourself.\n", | |
| "if 0 == 1:\n", | |
| "\n", | |
| " with codecs.open(unigram_sentences_filepath, 'w', encoding='utf_8') as f:\n", | |
| " for sentence in lemmatized_sentence_corpus(recall_txt_filepath):\n", | |
| " f.write(sentence + '\\n')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "If your data is organized like our `unigram_sentences_all` file now is — a large text file with one document/sentence per line — gensim's [**LineSentence**](https://radimrehurek.com/gensim/models/word2vec.html#gensim.models.word2vec.LineSentence) class provides a convenient iterator for working with other gensim components. It *streams* the documents/sentences from disk, so that you never have to hold the entire corpus in RAM at once. This allows you to scale your modeling pipeline up to potentially very large corpora." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 20, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "unigram_sentences = LineSentence(unigram_sentences_filepath)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Let's take a look at a few sample sentences in our new, transformed file." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 21, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "fda environmental sampling reveal the presence of microorganism and fungal growth in the clean room where sterile product be prepared\n", | |
| "\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "for unigram_sentence in it.islice(unigram_sentences, 23, 24):\n", | |
| " print(' '.join(unigram_sentence))\n", | |
| " print('')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Next, we'll learn a phrase model that will link individual words into two-word phrases. We'd expect words that together represent a specific concept, like \"`ice cream`\", to be linked together to form a new, single token: \"`ice_cream`\"." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 22, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "bigram_model_filepath = os.path.join(intermediate_directory, 'bigram_model_all')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 23, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "Wall time: 254 ms\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "%%time\n", | |
| "\n", | |
| "# this is a bit time consuming - make the if statement True\n", | |
| "# if you want to execute modeling yourself.\n", | |
| "if 0 == 1:\n", | |
| "\n", | |
| " bigram_model = Phrases(unigram_sentences)\n", | |
| "\n", | |
| " bigram_model.save(bigram_model_filepath)\n", | |
| " \n", | |
| "# load the finished model from disk\n", | |
| "bigram_model = Phrases.load(bigram_model_filepath)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Now that we have a trained phrase model for word pairs, let's apply it to the review sentences data and explore the results." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 24, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "bigram_sentences_filepath = os.path.join(intermediate_directory,\n", | |
| " 'bigram_sentences_all.txt')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 25, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "Wall time: 0 ns\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "%%time\n", | |
| "\n", | |
| "# this is a bit time consuming - make the if statement True\n", | |
| "# if you want to execute data prep yourself.\n", | |
| "if 0 == 1:\n", | |
| "\n", | |
| " with codecs.open(bigram_sentences_filepath, 'w', encoding='utf_8') as f:\n", | |
| " \n", | |
| " for unigram_sentence in unigram_sentences:\n", | |
| " \n", | |
| " bigram_sentence = ' '.join(bigram_model[unigram_sentence])\n", | |
| " \n", | |
| " f.write(bigram_sentence + '\\n')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 26, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "bigram_sentences = LineSentence(bigram_sentences_filepath)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 27, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "fda environmental_sampling reveal the presence of microorganism and fungal_growth in the clean_room where_sterile product be prepared\n", | |
| "\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "for bigram_sentence in it.islice(bigram_sentences, 23, 24):\n", | |
| " print(' '.join(bigram_sentence))\n", | |
| " print('')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Looks like the phrase modeling worked! We now see two-word phrases, such as \"`ice_cream`\" and \"`apple_pie`\", linked together in the text as a single token. Next, we'll train a _second-order_ phrase model. We'll apply the second-order phrase model on top of the already-transformed data, so that incomplete word combinations like \"`vanilla_ice cream`\" will become fully joined to \"`vanilla_ice_cream`\". No disrespect intended to [Vanilla Ice](https://www.youtube.com/watch?v=rog8ou-ZepE), of course." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 28, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "trigram_model_filepath = os.path.join(intermediate_directory,\n", | |
| " 'trigram_model_all')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 29, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "Wall time: 210 ms\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "%%time\n", | |
| "\n", | |
| "# this is a bit time consuming - make the if statement True\n", | |
| "# if you want to execute modeling yourself.\n", | |
| "if 0 == 1:\n", | |
| "\n", | |
| " trigram_model = Phrases(bigram_sentences)\n", | |
| "\n", | |
| " trigram_model.save(trigram_model_filepath)\n", | |
| " \n", | |
| "# load the finished model from disk\n", | |
| "trigram_model = Phrases.load(trigram_model_filepath)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "We'll apply our trained second-order phrase model to our first-order transformed sentences, write the results out to a new file, and explore a few of the second-order transformed sentences." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 30, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "trigram_sentences_filepath = os.path.join(intermediate_directory,\n", | |
| " 'trigram_sentences_all.txt')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 31, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "Wall time: 0 ns\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "%%time\n", | |
| "\n", | |
| "# this is a bit time consuming - make the if statement True\n", | |
| "# if you want to execute data prep yourself.\n", | |
| "if 0 == 1:\n", | |
| "\n", | |
| " with codecs.open(trigram_sentences_filepath, 'w', encoding='utf_8') as f:\n", | |
| " \n", | |
| " for bigram_sentence in bigram_sentences:\n", | |
| " \n", | |
| " trigram_sentence = ' '.join(trigram_model[bigram_sentence])\n", | |
| " \n", | |
| " f.write(trigram_sentence + '\\n')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 32, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "trigram_sentences = LineSentence(trigram_sentences_filepath)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 33, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "fda_environmental_sampling reveal the presence of microorganism and fungal_growth in the clean_room_where_sterile product be prepared\n", | |
| "\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "for trigram_sentence in it.islice(trigram_sentences, 23, 24):\n", | |
| " print(' '.join(trigram_sentence))\n", | |
| " print('')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Looks like the second-order phrase model was successful. We're now seeing three-word phrases, such as \"`vanilla_ice_cream`\" and \"`cinnamon_ice_cream`\"." | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "The final step of our text preparation process circles back to the complete text of the reviews. We're going to run the complete text of the reviews through a pipeline that applies our text normalization and phrase models.\n", | |
| "\n", | |
| "In addition, we'll remove stopwords at this point. _Stopwords_ are very common words, like _a_, _the_, _and_, and so on, that serve functional roles in natural language, but typically don't contribute to the overall meaning of text. Filtering stopwords is a common procedure that allows higher-level NLP modeling techniques to focus on the words that carry more semantic weight.\n", | |
| "\n", | |
| "Finally, we'll write the transformed text out to a new file, with one review per line." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 34, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "trigram_reviews_filepath = os.path.join(intermediate_directory,\n", | |
| " 'trigram_transformed_reviews_all.txt')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 35, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "Wall time: 0 ns\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| " %%time\n", | |
| "# %debug\n", | |
| "\n", | |
| "# this is a bit time consuming - make the if statement True\n", | |
| "# if you want to execute data prep yourself.\n", | |
| "if 0 == 1:\n", | |
| "\n", | |
| " with codecs.open(trigram_reviews_filepath, 'w', encoding='utf_8') as f:\n", | |
| " \n", | |
| " for parsed_reason in nlp.pipe(line_review(recall_txt_filepath),\n", | |
| " batch_size=10000, n_threads=4):\n", | |
| " \n", | |
| " # lemmatize the text, removing punctuation and whitespace\n", | |
| " unigram_review = [token.lemma_ for token in parsed_reason\n", | |
| " if not punct_space(token)]\n", | |
| " \n", | |
| " # apply the first-order and second-order phrase models\n", | |
| " bigram_review = bigram_model[unigram_review]\n", | |
| " trigram_review = trigram_model[bigram_review]\n", | |
| " \n", | |
| " # remove any remaining stopwords\n", | |
| " trigram_review = [term for term in trigram_review\n", | |
| " if term not in spacy.en.stop_words.STOP_WORDS]\n", | |
| " \n", | |
| " \n", | |
| " # write the transformed review as a line in the new file\n", | |
| " trigram_review = ' '.join(trigram_review)\n", | |
| " f.write(trigram_review + '\\n')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Let's preview the results. We'll grab one review from the file with the original, untransformed text, grab the same review from the file with the normalized and transformed text, and compare the two." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 37, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "Original:\n", | |
| "\n", | |
| "Lack of Assurance of Sterility: Franck's Lab Inc. initiated a recall of all Sterile Human Drugs distributed between 11/21/2011 and 05/21/2012 because FDA environmental sampling revealed the presence of microorganisms and fungal growth in the clean room where sterile products were prepared. \n", | |
| "\n", | |
| "----\n", | |
| "\n", | |
| "Transformed:\n", | |
| "\n", | |
| "lack assurance sterility franck_'s_lab_inc. initiate recall all_sterile_human drugs_distribute_between_11/21/2011 05/21/2012_because_fda environmental_sampling_reveal presence microorganism fungal_growth clean_room_where_sterile product prepared\n", | |
| "\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "print('Original:' + '\\n')\n", | |
| "\n", | |
| "for review in it.islice(line_review(recall_txt_filepath), 11, 12):\n", | |
| " print(review)\n", | |
| "\n", | |
| "print('----' + '\\n')\n", | |
| "print('Transformed:' + '\\n')\n", | |
| "\n", | |
| "with codecs.open(trigram_reviews_filepath, encoding='utf_8') as f:\n", | |
| " for review in it.islice(f, 11, 12):\n", | |
| " print(review)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "You can see that most of the grammatical structure has been scrubbed from the text — capitalization, articles/conjunctions, punctuation, spacing, etc. However, much of the general semantic *meaning* is still present. Also, multi-word concepts such as \"`friday_night`\" and \"`above_average`\" have been joined into single tokens, as expected. The review text is now ready for higher-level modeling. " | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "## Topic Modeling with Latent Dirichlet Allocation (_LDA_)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "*Topic modeling* is family of techniques that can be used to describe and summarize the documents in a corpus according to a set of latent \"topics\". For this demo, we'll be using [*Latent Dirichlet Allocation*](http://www.jmlr.org/papers/volume3/blei03a/blei03a.pdf) or LDA, a popular approach to topic modeling.\n", | |
| "\n", | |
| "In many conventional NLP applications, documents are represented a mixture of the individual tokens (words and phrases) they contain. In other words, a document is represented as a *vector* of token counts. There are two layers in this model — documents and tokens — and the size or dimensionality of the document vectors is the number of tokens in the corpus vocabulary. This approach has a number of disadvantages:\n", | |
| "* Document vectors tend to be large (one dimension for each token $\\Rightarrow$ lots of dimensions)\n", | |
| "* They also tend to be very sparse. Any given document only contains a small fraction of all tokens in the vocabulary, so most values in the document's token vector are 0.\n", | |
| "* The dimensions are fully indepedent from each other — there's no sense of connection between related tokens, such as _knife_ and _fork_.\n", | |
| "\n", | |
| "LDA injects a third layer into this conceptual model. Documents are represented as a mixture of a pre-defined number of *topics*, and the *topics* are represented as a mixture of the individual tokens in the vocabulary. The number of topics is a model hyperparameter selected by the practitioner. LDA makes a prior assumption that the (document, topic) and (topic, token) mixtures follow [*Dirichlet*](https://en.wikipedia.org/wiki/Dirichlet_distribution) probability distributions. This assumption encourages documents to consist mostly of a handful of topics, and topics to consist mostly of a modest set of the tokens." | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "LDA is fully unsupervised. The topics are \"discovered\" automatically from the data by trying to maximize the likelihood of observing the documents in your corpus, given the modeling assumptions. They are expected to capture some latent structure and organization within the documents, and often have a meaningful human interpretation for people familiar with the subject material.\n", | |
| "\n", | |
| "We'll again turn to gensim to assist with data preparation and modeling. In particular, gensim offers a high-performance parallelized implementation of LDA with its [**LdaMulticore**](https://radimrehurek.com/gensim/models/ldamulticore.html) class." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 38, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "from gensim.corpora import Dictionary, MmCorpus\n", | |
| "from gensim.models.ldamulticore import LdaMulticore\n", | |
| "\n", | |
| "import pyLDAvis\n", | |
| "import pyLDAvis.gensim\n", | |
| "import warnings\n", | |
| "import _pickle as pickle" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "The first step to creating an LDA model is to learn the full vocabulary of the corpus to be modeled. We'll use gensim's [**Dictionary**](https://radimrehurek.com/gensim/corpora/dictionary.html) class for this." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 39, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "trigram_dictionary_filepath = os.path.join(intermediate_directory,\n", | |
| " 'trigram_dict_all.dict')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 40, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "Wall time: 104 ms\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "%%time\n", | |
| "\n", | |
| "# this is a bit time consuming - make the if statement True\n", | |
| "# if you want to learn the dictionary yourself.\n", | |
| "if 0 == 1:\n", | |
| "\n", | |
| " trigram_reviews = LineSentence(trigram_reviews_filepath)\n", | |
| "\n", | |
| " # learn the dictionary by iterating over all of the reviews\n", | |
| " trigram_dictionary = Dictionary(trigram_reviews)\n", | |
| " \n", | |
| " # filter tokens that are very rare or too common from\n", | |
| " # the dictionary (filter_extremes) and reassign integer ids (compactify)\n", | |
| " trigram_dictionary.filter_extremes(no_below=10, no_above=0.4)\n", | |
| " trigram_dictionary.compactify()\n", | |
| "\n", | |
| " trigram_dictionary.save(trigram_dictionary_filepath)\n", | |
| " \n", | |
| "# load the finished dictionary from disk\n", | |
| "trigram_dictionary = Dictionary.load(trigram_dictionary_filepath)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Like many NLP techniques, LDA uses a simplifying assumption known as the [*bag-of-words* model](https://en.wikipedia.org/wiki/Bag-of-words_model). In the bag-of-words model, a document is represented by the counts of distinct terms that occur within it. Additional information, such as word order, is discarded. \n", | |
| "\n", | |
| "Using the gensim Dictionary we learned to generate a bag-of-words representation for each review. The `trigram_bow_generator` function implements this. We'll save the resulting bag-of-words reviews as a matrix.\n", | |
| "\n", | |
| "In the following code, \"bag-of-words\" is abbreviated as `bow`." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 41, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "trigram_bow_filepath = os.path.join(intermediate_directory,\n", | |
| " 'trigram_bow_corpus_all.mm')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 42, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "def trigram_bow_generator(filepath):\n", | |
| " \"\"\"\n", | |
| " generator function to read reviews from a file\n", | |
| " and yield a bag-of-words representation\n", | |
| " \"\"\"\n", | |
| " \n", | |
| " for reason in LineSentence(filepath):\n", | |
| " yield trigram_dictionary.doc2bow(reason)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 43, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "Wall time: 80 ms\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "%%time\n", | |
| "\n", | |
| "# this is a bit time consuming - make the if statement True\n", | |
| "# if you want to build the bag-of-words corpus yourself.\n", | |
| "if 0 == 1:\n", | |
| "\n", | |
| " # generate bag-of-words representations for\n", | |
| " # all reviews and save them as a matrix\n", | |
| " MmCorpus.serialize(trigram_bow_filepath,\n", | |
| " trigram_bow_generator(trigram_reviews_filepath))\n", | |
| " \n", | |
| "# load the finished bag-of-words corpus from disk\n", | |
| "trigram_bow_corpus = MmCorpus(trigram_bow_filepath)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "With the bag-of-words corpus, we're finally ready to learn our topic model from the reviews. We simply need to pass the bag-of-words matrix and Dictionary from our previous steps to `LdaMulticore` as inputs, along with the number of topics the model should learn. For this demo, we're asking for 50 topics." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 44, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "lda_model_filepath = os.path.join(intermediate_directory, 'lda_model_all')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 45, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "Wall time: 187 ms\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "%%time\n", | |
| "\n", | |
| "# this is a bit time consuming - make the if statement True\n", | |
| "# if you want to train the LDA model yourself.\n", | |
| "if 0 == 1:\n", | |
| "\n", | |
| " with warnings.catch_warnings():\n", | |
| " warnings.simplefilter('ignore')\n", | |
| " \n", | |
| " # workers => sets the parallelism, and should be\n", | |
| " # set to your number of physical cores minus one\n", | |
| " lda = LdaMulticore(trigram_bow_corpus,\n", | |
| " num_topics=5,\n", | |
| " id2word=trigram_dictionary,\n", | |
| " workers=1)\n", | |
| " \n", | |
| " lda.save(lda_model_filepath)\n", | |
| " \n", | |
| "# load the finished LDA model from disk\n", | |
| "lda = LdaMulticore.load(lda_model_filepath)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Our topic model is now trained and ready to use! Since each topic is represented as a mixture of tokens, you can manually inspect which tokens have been grouped together into which topics to try to understand the patterns the model has discovered in the data." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 46, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "def explore_topic(topic_number, topn=10):\n", | |
| " \"\"\"\n", | |
| " accept a user-supplied topic number and\n", | |
| " print out a formatted list of the top terms\n", | |
| " \"\"\"\n", | |
| " \n", | |
| " print('{:20} {}'.format(u'term', u'frequency') + '\\n')\n", | |
| "\n", | |
| " for term, frequency in lda.show_topic(topic_number, topn=10):\n", | |
| " print('{:20} {:.3f}'.format(term, round(frequency, 3)))" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 47, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "term frequency\n", | |
| "\n", | |
| "presence 0.048\n", | |
| "potency 0.040\n", | |
| "tablet 0.031\n", | |
| "specification_result 0.025\n", | |
| "product 0.023\n", | |
| "particulate_matter 0.023\n", | |
| "stability_data_do_not 0.020\n", | |
| "drug_package 0.020\n", | |
| "find 0.019\n", | |
| "support_expiry_potential_loss 0.018\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "explore_topic(topic_number=0)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 48, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "topic_names = {0: 'potency',\n", | |
| " 1: 'expiry',\n", | |
| " 2: 'penicillin',\n", | |
| " 3: 'tablets',\n", | |
| " 4: 'market without approved'}" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 49, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "topic_names_filepath = os.path.join(intermediate_directory, 'topic_names.pkl')\n", | |
| "\n", | |
| "with open(topic_names_filepath, 'wb') as f:\n", | |
| " pickle.dump(topic_names, f)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 50, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "LDAvis_data_filepath = os.path.join(intermediate_directory, 'ldavis_prepared')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 51, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "Wall time: 68 ms\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "%%time\n", | |
| "\n", | |
| "# this is a bit time consuming - make the if statement True\n", | |
| "# if you want to execute data prep yourself.\n", | |
| "if 0 == 1:\n", | |
| "\n", | |
| " LDAvis_prepared = pyLDAvis.gensim.prepare(lda, trigram_bow_corpus,\n", | |
| " trigram_dictionary)\n", | |
| "\n", | |
| " with open(LDAvis_data_filepath, 'wb') as f:\n", | |
| " pickle.dump(LDAvis_prepared, f)\n", | |
| " \n", | |
| "# load the pre-prepared pyLDAvis data from disk\n", | |
| "with open(LDAvis_data_filepath, \"rb\") as f:\n", | |
| " LDAvis_prepared = pickle.load(f)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "`pyLDAvis.display(...)` displays the topic model visualization in-line in the notebook." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 52, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "data": { | |
| "text/html": [ | |
| "\n", | |
| "<link rel=\"stylesheet\" type=\"text/css\" href=\"https://cdn.rawgit.com/bmabey/pyLDAvis/files/ldavis.v1.0.0.css\">\n", | |
| "\n", | |
| "\n", | |
| "<div id=\"ldavis_el1013626482925773521468873287\"></div>\n", | |
| "<script type=\"text/javascript\">\n", | |
| "\n", | |
| "var ldavis_el1013626482925773521468873287_data = {\"token.table\": {\"Freq\": [0.025287285551752813, 0.07586185665525844, 0.8850549943113484, 0.9170691719006715, 0.8710778090237391, 0.9710434702124949, 0.018673912888701825, 0.00746956515548073, 0.9085246822283154, 0.04129657646492343, 0.04129657646492343, 0.02693772099766765, 0.020203290748250738, 0.7879283391817787, 0.013468860498833826, 0.14815746548717207, 0.026037664761453024, 0.9633935961737619, 0.021537285783846075, 0.19383557205461466, 0.7214990737588435, 0.06461185735153822, 0.9522082809803416, 0.09659854213716955, 0.7727883370973564, 0.09659854213716955, 0.061005553283746856, 0.015251388320936714, 0.8159492751701142, 0.02287708248140507, 0.08388263576515192, 0.934825082076671, 0.022800611757967586, 0.04560122351593517, 0.025095827660698226, 0.8640134951754675, 0.0967981924055503, 0.010755354711727812, 0.01229185902770203, 0.9587650041607583, 0.01229185902770203, 0.02458371805540406, 0.037066497926241594, 0.9637289460822814, 0.02506432210339065, 0.8665094212886483, 0.09667667097022108, 0.010741852330024565, 0.009364403969470006, 0.9738980128248808, 0.009364403969470006, 0.009364403969470006, 0.9647101738991866, 0.004249824554621967, 0.03399859643697574, 0.9787169752176215, 0.003558970818973169, 0.007117941637946338, 0.003558970818973169, 0.003558970818973169, 0.10393474696241874, 0.8314779756993499, 0.064360048647034, 0.032180024323517, 0.22526017026461898, 0.6596904986320984, 0.04785738404111819, 0.023928692020559094, 0.19142953616447275, 0.6939320685962137, 0.023928692020559094, 0.9073729825168678, 0.05337488132452164, 0.05337488132452164, 0.11561239321230139, 0.20553314348853582, 0.012845821468033489, 0.6679827163377414, 0.868055064068153, 0.0510620625922443, 0.07659309388836645, 0.7178312614732243, 0.19747566831693117, 0.027675422129088165, 0.048143703078726285, 0.00864856941534005, 0.0665706113124524, 0.0665706113124524, 0.0665706113124524, 0.0665706113124524, 0.7322767244369764, 0.025287285551752813, 0.07586185665525844, 0.8850549943113484, 0.09271372740269557, 0.16224902295471724, 0.5496599553159809, 0.11589215925336947, 0.07946890920231049, 0.8706363137668621, 0.0842551271387286, 0.028085042379576198, 0.9253492324008544, 0.09531072112930132, 0.29864025953847745, 0.23509977878560992, 0.09531072112930132, 0.27957811531261717, 0.11478839675477154, 0.8494341359853094, 0.02295767935095431, 0.02295767935095431, 0.2918627243073504, 0.07888181738036497, 0.007888181738036498, 0.6152781755668468, 0.7062743136043406, 0.09664806396690975, 0.026020632606475705, 0.15612379563885423, 0.011151699688489588, 0.9509451780913356, 0.032791213037632265, 0.032791213037632265, 0.8336936000281754, 0.007791528972225938, 0.023374586916677813, 0.046749173833355626, 0.08960258318059829, 0.03953043174191613, 0.9408242754576039, 0.015812172696766453, 0.025119690982818587, 0.003588527283259798, 0.8648350752656113, 0.09689023664801455, 0.010765581849779394, 0.10336526281665284, 0.1624311272833116, 0.007383233058332346, 0.575892178549923, 0.14766466116664692, 0.10517032103906902, 0.6205048941305072, 0.26292580259767256, 0.010517032103906903, 0.018689822845106844, 0.9718707879455559, 0.018689822845106844, 0.11559772817781147, 0.2055070723161093, 0.01284419201975683, 0.6678979850273552, 0.010618694879936382, 0.13804303343917299, 0.010618694879936382, 0.8282582006350379, 0.010618694879936382, 0.11128365945280168, 0.011128365945280168, 0.8680125437318531, 0.17812419703405652, 0.22087400432223006, 0.3419984583053885, 0.18524916491541876, 0.07481216275430373, 0.9510230662244014, 0.06691059914480081, 0.06691059914480081, 0.8698377888824105, 0.025287285551752813, 0.07586185665525844, 0.8850549943113484, 0.9069135516736537, 0.07403375932029826, 0.018508439830074566, 0.868055064068153, 0.0510620625922443, 0.07659309388836645, 0.04442660071274655, 0.888532014254931, 0.04442660071274655, 0.04442660071274655, 0.13476918910882418, 0.10107689183161815, 0.72438439145993, 0.033692297277206046, 0.9869014457423825, 0.007591549582633711, 0.6460779416977954, 0.3230389708488977, 0.00929608549205461, 0.016268149611095566, 0.004648042746027305, 0.7425003488901964, 0.06750003171729059, 0.06750003171729059, 0.06750003171729059, 0.029749992452272295, 0.044624988678408445, 0.2826249282965868, 0.2528749358443145, 0.3941873999926079, 0.12263968945492004, 0.8584778261844404, 0.01635195859398934, 0.00817597929699467, 0.7945998819812402, 0.08828887577569336, 0.06621665683177003, 0.02207221894392334, 0.9732906018213383, 0.018789393857554793, 0.007515757543021917, 0.9191677186675825, 0.07070520912827558, 0.14133136991536105, 0.8479882194921663, 0.4542441816948739, 0.3726566344972509, 0.013230413059073997, 0.07056220298172798, 0.08820275372715998, 0.04215044103606922, 0.9273097027935228, 0.4495970623515219, 0.00449597062351522, 0.00449597062351522, 0.5395164748218263, 0.951700970790542, 0.02796000162471068, 0.9226800536154525, 0.02796000162471068, 0.9628340864004311, 0.01809838508271487, 0.014478708066171896, 0.003619677016542974, 0.010171828426742906, 0.010171828426742906, 0.010171828426742906, 0.02034365685348581, 0.9561518721138331, 0.028765972231986504, 0.014382986115993252, 0.21574479173989877, 0.7479152780316491, 0.017972428668250627, 0.9525387194172832, 0.017972428668250627, 0.007840833853823628, 0.9879450655817772, 0.055090650146544975, 0.055090650146544975, 0.8814504023447196, 0.908661350253221, 0.041302788647873685, 0.041302788647873685, 0.031440439823230776, 0.27248381180133335, 0.6497690896801026, 0.04192058643097436, 0.018694594916944807, 0.97211893568113, 0.018694594916944807, 0.1771290336423383, 0.5618031976130731, 0.008945910790017086, 0.25048550212047843, 0.0017891821580034173, 0.11561063484877263, 0.20553001750892913, 0.01284562609430807, 0.6679725569040197, 0.03825427831339644, 0.9181026795215146, 0.01912713915669822, 0.039533619362879135, 0.039533619362879135, 0.9092732453462201, 0.11486790995536807, 0.2010188424218941, 0.08934170774306405, 0.22654504463419814, 0.3669391568018702, 0.1961582468942004, 0.7646168399345362, 0.004003229528453069, 0.008006459056906138, 0.028022606699171483, 0.02508307181961939, 0.8635743297897533, 0.09674899130424623, 0.010749887922694025, 0.06697200988385313, 0.06697200988385313, 0.06697200988385313, 0.8036641186062375, 0.012360204503194734, 0.024720409006389468, 0.9146551332364103, 0.020600340838657888, 0.03296054534185262, 0.02791221998013107, 0.9490154793244563, 0.02791221998013107, 0.0962442520617253, 0.0962442520617253, 0.07699540164938024, 0.03849770082469012, 0.7122074652567671, 0.118582365306139, 0.04446838698980213, 0.014822795663267375, 0.014822795663267375, 0.8152537614797056, 0.023026948120335417, 0.003289564017190774, 0.8421283884008381, 0.11842430461886787, 0.013158256068763096, 0.08364186744422177, 0.8364186744422177, 0.06273140058316633, 0.02091046686105544, 0.6941504091055545, 0.05553203272844436, 0.24295264318694407, 0.006941504091055545, 0.9386108278038391, 0.03236589061392549, 0.03236589061392549, 0.9083182059177874, 0.02752479411872083, 0.02752479411872083, 0.02752479411872083, 0.13424665199344074, 0.04474888399781358, 0.8054799119606444, 0.056094196754554834, 0.12153742630153548, 0.6824793938470838, 0.06544322954698065, 0.07479226233940645, 0.9075227240096225, 0.03518377827314034, 0.09382340872837425, 0.19937474354779527, 0.07036755654628069, 0.5981242306433858, 0.1800701267612705, 0.060023375587090165, 0.060023375587090165, 0.6602571314579918, 0.130461042265656, 0.195691563398484, 0.65230521132828, 0.030728652921629837, 0.030728652921629837, 0.9064952611880802, 0.015364326460814919, 0.07953849030237271, 0.00994231128779659, 0.23861547090711813, 0.6760771675701681, 0.014620015105728622, 0.5738355928998484, 0.010965011329296465, 0.2924003021145724, 0.10965011329296466, 0.34759464813210617, 0.0993127566091732, 0.04256260997535994, 0.5107513197043193, 0.04821917108122678, 0.867945079462082, 0.04821917108122678, 0.04821917108122678, 0.960771823036405, 0.025966806028010943, 0.019541775084793264, 0.019541775084793264, 0.11725065050875959, 0.8012127784765238, 0.03908355016958653, 0.07355613380456014, 0.07355613380456014, 0.8091174718501615, 0.09624546717979279, 0.8662092046181351, 0.024061366794948198, 0.9725972346316365, 0.01263113291729398, 0.0315606996439763, 0.16569367313087557, 0.1499133233088874, 0.5838729434135616, 0.06706648674344963, 0.9519374164161721, 0.9492308219634603, 0.018612369058107064, 0.018747105892842335, 0.27183303544621384, 0.30932724723189853, 0.393689223749689, 0.010093212520543618, 0.023550829214601773, 0.7401689181731986, 0.20186425041087233, 0.023550829214601773, 0.04215044103606922, 0.9273097027935228, 0.7244091337914872, 0.06585537579922611, 0.06585537579922611, 0.06585537579922611, 0.06585537579922611, 0.779248367677424, 0.1714705509520599, 0.013167023109756782, 0.027531048320400542, 0.008678265231430607, 0.9398299737991166, 0.031327665793303885, 0.031327665793303885, 0.16126701941666519, 0.0537556731388884, 0.806335097083326, 0.42260229314728426, 0.40049694242881095, 0.05331290467396509, 0.03120755395549176, 0.09362266186647529, 0.03283586362616569, 0.9522400451588049, 0.01645210747267951, 0.9542222334154117, 0.01645210747267951, 0.01645210747267951, 0.12010993342877753, 0.7206596005726652, 0.12010993342877753, 0.3184350412170696, 0.5187409542407101, 0.08217678483021151, 0.0770407357783233, 0.00513604905188822, 0.7315222345009773, 0.18288055862524433, 0.00870859802977354, 0.07837738226796186, 0.00870859802977354, 0.04215044103606922, 0.9273097027935228, 0.047858297357728966, 0.023929148678864483, 0.19143318943091586, 0.6939453116870701, 0.023929148678864483, 0.04533345969863843, 0.24285781981413446, 0.019428625585130758, 0.6055254974032419, 0.08742881513308841, 0.1223236439954654, 0.011120331272315036, 0.8673858392405728, 0.013684245020723865, 0.02736849004144773, 0.9259672464023149, 0.01824566002763182, 0.01824566002763182, 0.09025451543862692, 0.09025451543862692, 0.8122906389476423, 0.05137855045388443, 0.06772627105284766, 0.7017842914269214, 0.054881633439376555, 0.12494329314921897, 0.04731083124570336, 0.02128987406056651, 0.6836415115004135, 0.11354599498968806, 0.13483586905025458, 0.02309595406096426, 0.01154797703048213, 0.5773988515241065, 0.12702774733530342, 0.265603471701089, 0.0168685796434513, 0.04217144910862825, 0.8771661414594676, 0.0337371592869026, 0.02530286946517695, 0.009449678563004786, 0.009449678563004786, 0.9638672134264883, 0.009449678563004786, 0.7517727228962107, 0.16342885280352404, 0.03268577056070481, 0.03268577056070481, 0.025906384997541415, 0.2479611135478964, 0.022205472855035498, 0.6143514156559821, 0.08882189142014199, 0.06429229883910095, 0.02143076627970032, 0.5786306895519086, 0.2285948403168034, 0.10001024263860149, 0.932822325416938, 0.0444201107341399, 0.035323005916963204, 0.0070646011833926416, 0.6004911005883745, 0.14835662485124546, 0.21193803550177923, 0.8474482614162128, 0.11770114741891845, 0.02354022948378369, 0.08796913242136803, 0.08796913242136803, 0.8796913242136802, 0.060860947986676316, 0.92508640939748, 0.012172189597335263, 0.9419120841880482, 0.032479727040967174, 0.032479727040967174, 0.8759987580556134, 0.04043071191025908, 0.08086142382051816, 0.034897568691853294, 0.9073367859881857, 0.06979513738370659, 0.10375201181449302, 0.14821715973499003, 0.007410857986749501, 0.5928686389399601, 0.14821715973499003, 0.030686344267739654, 0.9512766722999293, 0.015343172133869827, 0.07525790202654352, 0.0940723775331794, 0.07525790202654352, 0.7525790202654352, 0.06591752588219657, 0.922845362350752, 0.021972508627398857, 0.07088831553445107, 0.9038260230642512, 0.01772207888361277, 0.015546014830761777, 0.07773007415380888, 0.8239387860303742, 0.07773007415380888, 0.02873231821321078, 0.9481665010359557, 0.1127808071046346, 0.313671619759765, 0.07753680488443629, 0.010573200666059494, 0.4898916308607566, 0.38345703214820737, 0.046953922303862125, 0.5634470676463454, 0.09023615780482455, 0.09023615780482455, 0.812125420243421, 0.04865116327400725, 0.04865116327400725, 0.9243721022061379, 0.050422831678944374, 0.021332736479553388, 0.6458001134264798, 0.1473898156769143, 0.13381443791719852, 0.8315342797987751, 0.07059217356593847, 0.007266841396493666, 0.087202096757924, 0.0020762403989981904, 0.04977849023569283, 0.9258799183838866, 0.009955698047138566, 0.09755892828312147, 0.09755892828312147, 0.09755892828312147, 0.7804714262649718, 0.13364946892492163, 0.044549822974973875, 0.1484994099165796, 0.02969988198331592, 0.6533974036329502, 0.8949840015058175, 0.07181332538781608, 0.06104132657964367, 0.010771998808172412, 0.8581692383844022, 0.08480546565043948, 0.6431081145158327, 0.1837451755759522, 0.07773834351290286, 0.007067122137536623, 0.9912431433078739, 0.007595732898910911, 0.04276553487382613, 0.9337141780785372, 0.007127589145637689, 0.007127589145637689, 0.007127589145637689, 0.08849595703730372, 0.11061994629662965, 0.06637196777797778, 0.7300916455577556, 0.052973907831875465, 0.0397304308739066, 0.5363608167977391, 0.09270433870578207, 0.28473475459633063, 0.1193474083942662, 0.2688595463826876, 0.05639492924124667, 0.17311931767080374, 0.38164940486518095, 0.03387344780867308, 0.9230514527863415, 0.03387344780867308, 0.00846836195216827, 0.03410180487002054, 0.9093814632005477, 0.011367268290006846, 0.03410180487002054, 0.011367268290006846, 0.9171351206854794, 0.04663398918739726, 0.015544663062465753, 0.007772331531232877, 0.007772331531232877, 0.9768325344284536, 0.011628958743195876, 0.38345703214820737, 0.046953922303862125, 0.5634470676463454, 0.32124034913552213, 0.4050970055339999, 0.07345338699565913, 0.15604904104228012, 0.04381983172702412, 0.05687171469145639, 0.008124530670208055, 0.6987096376378927, 0.040622653351040275, 0.19498873608499334, 0.09664968564470205, 0.8698471708023185, 0.024162421411175514, 0.39636956305317506, 0.5637256007867378, 0.007046570009834223, 0.03347120754671256, 0.9231719860530064, 0.03692687944212026, 0.04635425455237729, 0.6158493819101555, 0.32447978186664106, 0.006622036364625327, 0.7834062222427162, 0.060262017095593555, 0.060262017095593555, 0.060262017095593555, 0.09416501680130175, 0.8474851512117157, 0.0995557458167954, 0.6071122713649219, 0.03822229526894823, 0.173333664591742, 0.08177793406379621, 0.8676974755725869, 0.019795379670857496, 0.013196919780571664, 0.029693069506286245, 0.07258305879314415, 0.03402032727935019, 0.620870972848141, 0.02126270454959387, 0.3231931091538268, 0.15202250691271363, 0.8209215373286536, 0.03353829605338779, 0.939072289494858, 0.03353829605338779, 0.9760102285286548, 0.01635771332729589, 0.00545257110909863, 0.08584441788288705, 0.08584441788288705, 0.08584441788288705, 0.7725997609459834, 0.2677489990373211, 0.3321443279197147, 0.2779166825450674, 0.023724594851408196, 0.1016768350774637, 0.29115031752375586, 0.08655820250706256, 0.007868927500642052, 0.6059074175494379, 0.06186517241048698, 0.8042472413363307, 0.12373034482097396, 0.016255163210874307, 0.08127581605437155, 0.8127581605437154, 0.08127581605437155, 0.031008052925124666, 0.031008052925124666, 0.93024158775374, 0.12736395839762105, 0.859706719183942, 0.007960247399851315, 0.09022553375736592, 0.09022553375736592, 0.8120298038162932, 0.0617670178774345, 0.0617670178774345, 0.741204214529214, 0.123534035754869, 0.025146483226097104, 0.8621651391804721, 0.10058593290438841, 0.010777064239755902, 0.08072637868280774, 0.09635083907302859, 0.263011749902051, 0.36717481917019007, 0.19530575487776067, 0.004732148197777908, 0.014196444593333725, 0.2224109652955617, 0.042589333780001175, 0.7192865260622421, 0.04072329170624159, 0.013574430568747197, 0.09502101398123038, 0.7058703895748543, 0.16289316682496635, 0.513831078279984, 0.01875295906131328, 0.0037505918122626566, 0.011251775436787969, 0.4538216092837814, 0.039392708447962034, 0.9454250027510888, 0.3477119523261312, 0.04090728850895661, 0.6136093276343492, 0.4225380022375716, 0.5671906516522357, 0.003806648668806951, 0.8757106677311117, 0.004759297107234303, 0.019037188428937212, 0.09994523925192036, 0.04306978819292141, 0.021534894096460706, 0.8183259756655069, 0.10767447048230354, 0.8791768001534532, 0.02313623158298561, 0.06940869474895683, 0.0038539234269604886, 0.5626728203362313, 0.050101004550486355, 0.10790985595489369, 0.2736285633141947, 0.05244587731427367, 0.10489175462854734, 0.12237371373330523, 0.10489175462854734, 0.5943866095617683, 0.05225847582217905, 0.13064618955544763, 0.026129237911089526, 0.07838771373326858, 0.7054894235994172, 0.45923001923281487, 0.004783646033675155, 0.004783646033675155, 0.526201063704267, 0.9596297956812535, 0.025935940423817663, 0.5676058721152761, 0.02365024467146984, 0.003941707445244973, 0.02365024467146984, 0.3823456221887624, 0.03314697208728716, 0.05578392863470278, 0.6710240690841058, 0.08893090072198993, 0.15037406849354662, 0.2874873396724207, 0.020534809976601478, 0.10267404988300738, 0.5955094893214428, 0.023573872094588024, 0.05500570155403872, 0.19644893412156686, 0.2514546356756056, 0.47933539925662316, 0.2917746169474596, 0.08674380503843394, 0.007885800458039448, 0.6150924357270771, 0.11559580229859055, 0.20550364853082767, 0.01284397803317673, 0.6678868577251899, 0.1120534001642519, 0.028013350041062975, 0.8123871511908263, 0.05602670008212595, 0.05533455873925004, 0.9130202191976257, 0.02766727936962502, 0.02988536647942122, 0.23908293183536977, 0.7172487955061093, 0.47752976217727106, 0.1121714206456677, 0.016024488663666812, 0.35894854606613663, 0.03525387506006699, 0.09147771214680496, 0.7489737682019656, 0.1143471401835062, 0.02286942803670124, 0.017152071027525927, 0.1770200592591106, 0.5596694802838548, 0.010728488439946096, 0.2503313969320756, 0.0017880814066576827, 0.06461144969798188, 0.06461144969798188, 0.8399488460737645, 0.00970324771811111, 0.174658458926, 0.00970324771811111, 0.7568533220126665, 0.04851623859055555, 0.9759312362783739, 0.018072800671821737, 0.018072800671821737, 0.00902302919596078, 0.8075611130384899, 0.00451151459798039, 0.17820482662022544, 0.9915280731831905, 0.007597916269602992], \"Topic\": [3, 4, 5, 3, 3, 1, 2, 3, 2, 3, 5, 1, 2, 3, 4, 5, 3, 5, 1, 2, 3, 5, 3, 3, 4, 5, 1, 2, 3, 4, 5, 2, 3, 4, 1, 3, 4, 5, 1, 2, 3, 5, 3, 4, 1, 3, 4, 5, 1, 2, 3, 4, 2, 3, 5, 1, 2, 3, 4, 5, 3, 4, 2, 3, 4, 5, 1, 2, 3, 4, 5, 3, 4, 5, 1, 2, 3, 4, 1, 3, 4, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 3, 4, 5, 1, 2, 3, 4, 5, 2, 3, 4, 3, 1, 2, 3, 4, 5, 1, 2, 3, 5, 1, 2, 3, 4, 1, 2, 3, 4, 5, 1, 2, 3, 1, 2, 3, 4, 5, 1, 2, 3, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 5, 1, 2, 3, 1, 2, 3, 4, 1, 2, 3, 4, 5, 2, 3, 4, 1, 2, 3, 4, 5, 3, 2, 3, 5, 3, 4, 5, 2, 3, 4, 1, 3, 4, 1, 2, 3, 5, 2, 3, 4, 5, 1, 3, 1, 2, 3, 4, 5, 1, 2, 3, 4, 1, 2, 3, 4, 5, 2, 3, 4, 5, 1, 2, 3, 5, 1, 2, 3, 1, 3, 1, 3, 1, 2, 3, 4, 5, 3, 4, 1, 2, 3, 5, 3, 2, 3, 4, 1, 2, 3, 4, 1, 2, 3, 4, 5, 1, 3, 4, 5, 1, 3, 5, 1, 3, 2, 3, 4, 2, 3, 5, 1, 3, 4, 5, 1, 2, 3, 1, 2, 3, 4, 5, 1, 2, 3, 4, 1, 3, 5, 1, 3, 4, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 3, 4, 5, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 1, 2, 3, 4, 1, 2, 3, 1, 2, 3, 4, 1, 3, 4, 1, 2, 3, 4, 5, 3, 1, 2, 3, 4, 5, 2, 3, 4, 5, 3, 4, 5, 1, 2, 3, 5, 1, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 1, 2, 3, 4, 1, 3, 1, 2, 3, 4, 5, 3, 4, 5, 1, 3, 4, 3, 5, 1, 2, 3, 4, 5, 3, 2, 3, 2, 3, 4, 5, 1, 2, 3, 4, 5, 3, 4, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 2, 3, 5, 2, 3, 4, 1, 2, 3, 4, 5, 1, 3, 1, 3, 4, 5, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 3, 4, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 2, 3, 4, 1, 2, 3, 4, 5, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 5, 1, 2, 3, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 2, 3, 1, 2, 3, 4, 5, 1, 2, 3, 2, 3, 4, 1, 2, 3, 1, 2, 3, 2, 3, 5, 1, 3, 4, 1, 2, 3, 4, 5, 1, 2, 3, 1, 3, 4, 5, 1, 3, 4, 1, 3, 4, 2, 3, 4, 5, 3, 4, 1, 2, 3, 4, 5, 1, 3, 4, 3, 4, 5, 1, 3, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 1, 3, 4, 5, 1, 2, 3, 4, 5, 3, 1, 2, 3, 5, 1, 2, 3, 4, 5, 1, 3, 1, 2, 3, 4, 5, 1, 2, 3, 4, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 3, 1, 3, 4, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 3, 4, 1, 2, 3, 4, 2, 3, 1, 2, 3, 5, 1, 3, 4, 5, 1, 3, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 3, 5, 1, 2, 3, 1, 2, 3, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 3, 4, 5, 1, 2, 3, 5, 1, 3, 4, 1, 2, 3, 3, 4, 5, 1, 3, 4, 5, 1, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 3, 5, 3, 4, 5, 1, 2, 3, 1, 3, 4, 5, 2, 3, 4, 5, 2, 3, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 5, 1, 3, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 1, 2, 3, 4, 1, 3, 4, 5, 1, 2, 3, 2, 3, 4, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 3, 4, 5, 1, 2, 3, 4, 5, 2, 3, 4, 1, 2, 3, 4, 1, 3], \"Term\": [\"/capsule\", \"/capsule\", \"/capsule\", \"0.125\", \"00904052260_pedigree\", \"02/12/15\", \"02/12/15\", \"02/12/15\", \"05/21/2012\", \"05/21/2012\", \"05/21/2012\", \"10\", \"10\", \"10\", \"10\", \"10\", \"18_month_stability_time\", \"18_month_stability_time\", \"2\", \"2\", \"2\", \"2\", \"20%\", \"36_month\", \"36_month\", \"36_month\", \"500\", \"500\", \"500\", \"500\", \"500\", \"6_month\", \"6_month\", \"6_month\", \"80mg/ml_10_ml_vial\", \"80mg/ml_10_ml_vial\", \"80mg/ml_10_ml_vial\", \"80mg/ml_10_ml_vial\", \"active_ingredient_that\", \"active_ingredient_that\", \"active_ingredient_that\", \"active_ingredient_that\", \"active_sunscreen_ingredient\", \"active_sunscreen_ingredient\", \"adverse_reaction\", \"adverse_reaction\", \"adverse_reaction\", \"adverse_reaction\", \"affect\", \"affect\", \"affect\", \"affect\", \"all_sterile_human\", \"all_sterile_human\", \"all_sterile_human\", \"also_cgmp_deviation\", \"also_cgmp_deviation\", \"also_cgmp_deviation\", \"also_cgmp_deviation\", \"also_cgmp_deviation\", \"an_analogue\", \"an_analogue\", \"and/or_exp\", \"and/or_exp\", \"and/or_exp\", \"and/or_exp\", \"approve_nda/anda\", \"approve_nda/anda\", \"approve_nda/anda\", \"approve_nda/anda\", \"approve_nda/anda\", \"ascorbic_acid\", \"ascorbic_acid\", \"ascorbic_acid\", \"aseptic_practice\", \"aseptic_practice\", \"aseptic_practice\", \"aseptic_practice\", \"assay_or\", \"assay_or\", \"assay_or\", \"assurance\", \"assurance\", \"assurance\", \"assurance\", \"assurance\", \"black\", \"black\", \"black\", \"black\", \"black\", \"blister_cavity_may\", \"blister_cavity_may\", \"blister_cavity_may\", \"bottle\", \"bottle\", \"bottle\", \"bottle\", \"bottle\", \"broken\", \"broken\", \"broken\", \"by_c._difficile_discover\", \"capsule\", \"capsule\", \"capsule\", \"capsule\", \"capsule\", \"certain\", \"certain\", \"certain\", \"certain\", \"certain_quality_control_procedure\", \"certain_quality_control_procedure\", \"certain_quality_control_procedure\", \"certain_quality_control_procedure\", \"cgmp_deviation\", \"cgmp_deviation\", \"cgmp_deviation\", \"cgmp_deviation\", \"cgmp_deviation\", \"cgmp_deviation_pharmaceutical\", \"cgmp_deviation_pharmaceutical\", \"cgmp_deviation_pharmaceutical\", \"compound\", \"compound\", \"compound\", \"compound\", \"compound\", \"compound_by\", \"compound_by\", \"compound_by\", \"compound_preservative_free_methylprednisolone\", \"compound_preservative_free_methylprednisolone\", \"compound_preservative_free_methylprednisolone\", \"compound_preservative_free_methylprednisolone\", \"compound_preservative_free_methylprednisolone\", \"compound_sterile_preparation\", \"compound_sterile_preparation\", \"compound_sterile_preparation\", \"compound_sterile_preparation\", \"compound_sterile_preparation\", \"concern_associate\", \"concern_associate\", \"concern_associate\", \"concern_associate\", \"concern_regard_quality_control\", \"concern_regard_quality_control\", \"concern_regard_quality_control\", \"condition_potentially\", \"condition_potentially\", \"condition_potentially\", \"condition_potentially\", \"conduct\", \"conduct\", \"conduct\", \"conduct\", \"conduct\", \"connection\", \"connection\", \"connection\", \"contain\", \"contain\", \"contain\", \"contain\", \"contain\", \"contain_62%_ethyl_alcohol\", \"contain_foreign_substance\", \"contain_foreign_substance\", \"contain_foreign_substance\", \"contain_more_than_one\", \"contain_more_than_one\", \"contain_more_than_one\", \"contain_undeclared_sibutramine\", \"contain_undeclared_sibutramine\", \"contain_undeclared_sibutramine\", \"content_uniformity_failure\", \"content_uniformity_failure\", \"content_uniformity_failure\", \"control\", \"control\", \"control\", \"control\", \"correct\", \"correct\", \"correct\", \"correct\", \"could_introduce\", \"could_introduce\", \"cross_contamination\", \"cross_contamination\", \"cross_contamination\", \"cross_contamination\", \"cross_contamination\", \"customer\", \"customer\", \"customer\", \"customer\", \"date\", \"date\", \"date\", \"date\", \"date\", \"defective_container\", \"defective_container\", \"defective_container\", \"defective_container\", \"distribute\", \"distribute\", \"distribute\", \"distribute\", \"distribute_between_01/05/12\", \"distribute_between_01/05/12\", \"distribute_between_01/05/12\", \"distribute_by_this\", \"distribute_by_this\", \"documentation\", \"documentation\", \"drug\", \"drug\", \"drug\", \"drug\", \"drug\", \"drug/disease_claim_make_them\", \"drug/disease_claim_make_them\", \"drug_package\", \"drug_package\", \"drug_package\", \"drug_package\", \"ethyl_alcohol_content\", \"exp_6/26/2014\", \"exp_6/26/2014\", \"exp_6/26/2014\", \"facility\", \"facility\", \"facility\", \"facility\", \"fail_impurities/degradation_specification\", \"fail_impurities/degradation_specification\", \"fail_impurities/degradation_specification\", \"fail_impurities/degradation_specification\", \"fail_impurities/degradation_specification\", \"fail_impurities/degradation_specifications_out\", \"fail_impurities/degradation_specifications_out\", \"fail_impurities/degradation_specifications_out\", \"fail_impurities/degradation_specifications_out\", \"fail_impurity/degradation_specification\", \"fail_impurity/degradation_specification\", \"fail_impurity/degradation_specification\", \"failed_dissolution_specification\", \"failed_dissolution_specification\", \"failed_lozenge_specifications\", \"failed_lozenge_specifications\", \"failed_lozenge_specifications\", \"fda_environmental_sampling\", \"fda_environmental_sampling\", \"fda_environmental_sampling\", \"fda_inspection\", \"fda_inspection\", \"fda_inspection\", \"fda_inspection\", \"fda_inspection_finding_result\", \"fda_inspection_finding_result\", \"fda_inspection_finding_result\", \"fda_inspection_identify_gmp\", \"fda_inspection_identify_gmp\", \"fda_inspection_identify_gmp\", \"fda_inspection_identify_gmp\", \"fda_inspection_identify_gmp\", \"fda_inspection_reveal_poor\", \"fda_inspection_reveal_poor\", \"fda_inspection_reveal_poor\", \"fda_inspection_reveal_poor\", \"fda_inspectional_finding_result\", \"fda_inspectional_finding_result\", \"fda_inspectional_finding_result\", \"fda_sampling_confirm\", \"fda_sampling_confirm\", \"fda_sampling_confirm\", \"find\", \"find\", \"find\", \"find\", \"find\", \"firm\", \"firm\", \"firm\", \"firm\", \"firm\", \"firm_receive_seven_report\", \"firm_receive_seven_report\", \"firm_receive_seven_report\", \"firm_receive_seven_report\", \"float\", \"float\", \"float\", \"float\", \"follow_drug\", \"follow_drug\", \"follow_drug\", \"follow_drug\", \"follow_drug\", \"for_safety_reason\", \"for_safety_reason\", \"for_safety_reason\", \"foreign_substance\", \"foreign_substance\", \"foreign_substance\", \"foreign_substance\", \"foreign_substance\", \"foreign_tablets/capsule\", \"foreign_tablets/capsule\", \"foreign_tablets/capsule\", \"foreign_tablets/capsule\", \"foreign_tablets/capsule\", \"form\", \"form\", \"form\", \"form\", \"form\", \"glass\", \"glass\", \"glass\", \"glass\", \"glass_particulate\", \"glass_particulate\", \"glass_particulate\", \"glass_particulate\", \"good_manufacturing_practices\", \"good_manufacturing_practices\", \"good_manufacturing_practices\", \"guarantee\", \"guarantee\", \"guarantee\", \"guarantee\", \"have_an\", \"have_an\", \"have_an\", \"hcl\", \"hcl\", \"hcl\", \"hcl\", \"hcl\", \"hyoscyamine_sulfate_sl\", \"identify_as\", \"identify_as\", \"identify_as\", \"identify_as\", \"identify_as\", \"identify_during\", \"identify_during\", \"identify_during\", \"identify_during\", \"illegible\", \"illegible\", \"illegible\", \"impact\", \"impact\", \"impact\", \"impact\", \"impurity\", \"impurity\", \"impurity\", \"impurity\", \"initiate\", \"initiate\", \"initiate\", \"initiate\", \"initiate\", \"injectable_drug\", \"injectable_drug\", \"injectable_drug\", \"injectable_drug\", \"inspection\", \"inspection\", \"inspection\", \"inspection\", \"inspection_observation_associate\", \"inspection_observation_associate\", \"instead\", \"instead\", \"instead\", \"instead\", \"instead\", \"interval\", \"interval\", \"interval\", \"iv_bag_leak\", \"iv_bag_leak\", \"iv_bag_leak\", \"know_impurity\", \"know_impurity\", \"label\", \"label\", \"label\", \"label\", \"label\", \"label_state\", \"labeling:label_mixup\", \"labeling:label_mixup\", \"labeling_incorrect_or_missing\", \"labeling_incorrect_or_missing\", \"labeling_incorrect_or_missing\", \"labeling_incorrect_or_missing\", \"labeling_label_mixup\", \"labeling_label_mixup\", \"labeling_label_mixup\", \"labeling_label_mixup\", \"labeling_label_mixup\", \"labeling_that_bear\", \"labeling_that_bear\", \"laboratory\", \"laboratory\", \"laboratory\", \"laboratory\", \"laboratory\", \"lack\", \"lack\", \"lack\", \"lack\", \"lack\", \"leakage\", \"leakage\", \"leakage\", \"less_than\", \"less_than\", \"less_than\", \"lot\", \"lot\", \"lot\", \"lot\", \"lot\", \"lot_and/or_expiration\", \"lot_and/or_expiration\", \"lozenge\", \"lozenge\", \"lozenge\", \"lozenge\", \"male_erectile\", \"male_erectile\", \"male_erectile\", \"manufacture\", \"manufacture\", \"manufacture\", \"manufacture\", \"manufacture\", \"manufacturer\", \"manufacturer\", \"manufacturer\", \"manufacturer\", \"manufacturer\", \"mark_as_dietary_supplement\", \"mark_as_dietary_supplement\", \"market_without\", \"market_without\", \"market_without\", \"market_without\", \"market_without\", \"market_without_an_approved\", \"market_without_an_approved\", \"market_without_an_approved\", \"market_without_an_approved\", \"market_without_an_approved\", \"martin_avenue_pharmacy_inc.\", \"martin_avenue_pharmacy_inc.\", \"martin_avenue_pharmacy_inc.\", \"may_have_potentially\", \"may_have_potentially\", \"may_have_potentially\", \"may_have_potentially\", \"may_have_potentially\", \"medical_inc.\", \"medical_inc.\", \"medical_inc.\", \"mg\", \"mg\", \"mg\", \"mg\", \"mg\", \"mg_ndc\", \"mg_ndc\", \"mg_ndc\", \"mg_ndc\", \"mg_ndc\", \"microbial_contamination\", \"microbial_contamination\", \"microbial_contamination\", \"microbial_contamination\", \"microbial_contamination\", \"mislabeled_as\", \"mislabeled_as\", \"mislabeled_as\", \"mislabeled_as\", \"mislabeled_as\", \"mislabeled_as_one\", \"mislabeled_as_one\", \"mislabeled_as_one\", \"mislabeled_as_one\", \"ml\", \"ml\", \"ml\", \"ml\", \"nda/anda\", \"nda/anda\", \"nda/anda\", \"nda/anda\", \"nda/anda\", \"ndc\", \"ndc\", \"ndc\", \"ndc\", \"ndc\", \"neck\", \"neck\", \"non_sterile\", \"non_sterile\", \"non_sterile\", \"non_sterile\", \"non_sterile\", \"not_assure\", \"not_assure\", \"not_assure\", \"not_elsewhere_classify\", \"not_elsewhere_classify\", \"not_elsewhere_classify\", \"not_expire_due\", \"not_expire_due\", \"not_expire_due\", \"not_manufacture_accord\", \"not_manufacture_accord\", \"not_manufacture_accord\", \"number\", \"number\", \"number\", \"objectionable_condition_observe_during\", \"objectionable_condition_observe_during\", \"objectionable_condition_observe_during\", \"observation_associate\", \"observation_associate\", \"observation_associate\", \"observation_associate\", \"observation_associate\", \"observe_during\", \"observe_during\", \"observe_during\", \"obtain\", \"obtain\", \"obtain\", \"obtain\", \"overly_thick_overly_soft\", \"overly_thick_overly_soft\", \"overly_thick_overly_soft\", \"overwrap\", \"overwrap\", \"overwrap\", \"package_insert\", \"package_insert\", \"package_insert\", \"package_insert\", \"panel\", \"panel\", \"particulate_matter\", \"particulate_matter\", \"particulate_matter\", \"particulate_matter\", \"particulate_matter\", \"particulate_matter_api_contaminate\", \"particulate_matter_api_contaminate\", \"particulate_matter_api_contaminate\", \"particulate_matter_b._braun\", \"particulate_matter_b._braun\", \"particulate_matter_b._braun\", \"particulate_matter_confirm_customer\", \"particulate_matter_confirm_customer\", \"particulate_matter_confirm_customer\", \"pedigree\", \"pedigree\", \"pedigree\", \"pedigree\", \"pedigree\", \"penicillin\", \"penicillin\", \"penicillin\", \"penicillin\", \"penicillin\", \"pharmacy_that\", \"pharmacy_that\", \"pharmacy_that\", \"plastic\", \"plastic\", \"plastic\", \"plastic\", \"point\", \"point\", \"point\", \"point\", \"point\", \"possible_microbial_contamination\", \"potency\", \"potency\", \"potency\", \"potency\", \"potential\", \"potential\", \"potential\", \"potential\", \"potential\", \"potential_for_cross_contamination\", \"potential_for_cross_contamination\", \"potential_risk\", \"potential_risk\", \"potential_risk\", \"potential_risk\", \"potential_risk\", \"potentially_contaminate\", \"potentially_contaminate\", \"potentially_contaminate\", \"potentially_contaminate\", \"potentially_mislabeled_as\", \"potentially_mislabeled_as\", \"potentially_mislabeled_as\", \"potentially_mislabeled_as\", \"potentially_mislabeled_as\", \"presence\", \"presence\", \"presence\", \"presence\", \"presence\", \"present\", \"present\", \"present\", \"present\", \"process\", \"process\", \"process\", \"process\", \"process\", \"processing_controls\", \"processing_controls\", \"processing_controls\", \"processing_controls\", \"processing_controls\", \"produce\", \"produce\", \"produce_sterile\", \"produce_sterile\", \"produce_sterile\", \"product\", \"product\", \"product\", \"product\", \"product\", \"products\", \"products\", \"products\", \"products\", \"products\", \"puncture_through\", \"puncture_through\", \"puncture_through\", \"quality\", \"quality\", \"quality\", \"quality\", \"quality_control_procedure\", \"quality_control_procedure\", \"quality_control_procedure_that\", \"quality_control_procedure_that\", \"quality_control_procedure_that\", \"quality_control_procedure_that\", \"quality_control_process\", \"quality_control_process\", \"quality_control_process\", \"quality_control_process\", \"quetiapine_fumarate\", \"quetiapine_fumarate\", \"recall\", \"recall\", \"recall\", \"recall\", \"recall\", \"recall_because_they\", \"recall_because_they\", \"recall_because_they\", \"recall_because_they\", \"recall_because_they\", \"recent_fda_inspection\", \"recent_fda_inspection\", \"recent_fda_inspection\", \"recent_fda_inspection\", \"related\", \"related\", \"remove_from\", \"remove_from\", \"remove_from\", \"repackaged\", \"repackaged\", \"repackaged\", \"reserve_sample_unit\", \"reserve_sample_unit\", \"reserve_sample_unit\", \"reserve_sample_unit\", \"result\", \"result\", \"result\", \"result\", \"result\", \"risk\", \"risk\", \"risk\", \"risk\", \"room_temperature\", \"room_temperature\", \"room_temperature\", \"rubber\", \"rubber\", \"rubber\", \"rubber\", \"salicylic_acid\", \"salicylic_acid\", \"salicylic_acid\", \"select_sterile\", \"select_sterile\", \"select_sterile\", \"several_injectable\", \"several_injectable\", \"several_injectable\", \"sildenafil\", \"sildenafil\", \"sildenafil\", \"sildenafil\", \"skin_abscess_potentially_link\", \"skin_abscess_potentially_link\", \"skin_abscess_potentially_link\", \"skin_abscess_potentially_link\", \"specification\", \"specification\", \"specification\", \"specification\", \"specification\", \"specification_result\", \"specification_result\", \"specification_result\", \"specification_result\", \"specification_result\", \"stability\", \"stability\", \"stability\", \"stability\", \"stability\", \"stability_data_do_not\", \"stability_data_do_not\", \"stability_data_do_not\", \"stability_data_do_not\", \"stability_data_do_not\", \"stability_datum_do_not\", \"stability_datum_do_not\", \"stability_time_point\", \"stability_time_point\", \"stability_time_point\", \"sterile\", \"sterile\", \"sterile\", \"store\", \"store\", \"store\", \"store\", \"strength\", \"strength\", \"strength\", \"strength\", \"subject\", \"subject\", \"subject\", \"subpotent_drug\", \"subpotent_drug\", \"subpotent_drug\", \"subpotent_drug\", \"subpotent_drug\", \"superpotent_drug\", \"superpotent_drug\", \"superpotent_drug\", \"superpotent_drug\", \"superpotent_drug\", \"support_expiry\", \"support_expiry\", \"support_expiry\", \"support_expiry\", \"support_expiry\", \"support_expiry_potential_loss\", \"support_expiry_potential_loss\", \"support_expiry_potential_loss\", \"support_expiry_potential_loss\", \"support_expiry_recent\", \"support_expiry_recent\", \"syringe\", \"syringe\", \"syringe\", \"syringe\", \"syringe\", \"tablet\", \"tablet\", \"tablet\", \"tablet\", \"tablet\", \"temperature_abuse\", \"temperature_abuse\", \"temperature_abuse\", \"temperature_abuse\", \"test\", \"test\", \"test\", \"test\", \"test\", \"that_present\", \"that_present\", \"that_present\", \"that_present\", \"their_compound\", \"their_compound\", \"their_compound\", \"their_compound\", \"total_impurity\", \"total_impurity\", \"total_impurity\", \"total_impurity\", \"u.s._market\", \"u.s._market\", \"u.s._market\", \"unapproved_drug\", \"unapproved_drug\", \"unapproved_drug\", \"use\", \"use\", \"use\", \"use\", \"use\", \"vial\", \"vial\", \"vial\", \"vial\", \"vial\", \"violation_potentially_impact\", \"violation_potentially_impact\", \"violation_potentially_impact\", \"violation_potentially_impact\", \"violation_potentially_impact\", \"visible_particulate_matter\", \"visible_particulate_matter\", \"visible_particulate_matter\", \"voluntary\", \"voluntary\", \"voluntary\", \"voluntary\", \"voluntary\", \"withdraw_from\", \"withdraw_from\", \"withdraw_from\", \"within_expiry\", \"within_expiry\", \"within_expiry\", \"within_expiry\", \"without_adequate_separation_which\", \"without_adequate_separation_which\"]}, \"mdsDat\": {\"cluster\": [1, 1, 1, 1, 1], \"Freq\": [29.14085865023037, 25.223213794128608, 21.095014648739994, 13.663551886254787, 10.877361020646234], \"topics\": [1, 2, 3, 4, 5], \"y\": [-0.07302220687329498, 0.04909584417633757, -0.1403934838412857, 0.02649834475845766, 0.13782150177978547], \"x\": [-0.23486701075716332, -0.1260325021127151, 0.21245919891683787, 0.014311624720131175, 0.1341286892329094]}, \"tinfo\": {\"logprob\": [30.0, 29.0, 28.0, 27.0, 26.0, 25.0, 24.0, 23.0, 22.0, 21.0, 20.0, 19.0, 18.0, 17.0, 16.0, 15.0, 14.0, 13.0, 12.0, 11.0, 10.0, 9.0, 8.0, 7.0, 6.0, 5.0, 4.0, 3.0, 2.0, 1.0, -4.1268, -4.1269, -4.1272, -5.2535, -4.0713, -3.4044, -4.1271, -4.1337, -4.1077, -6.0803, -6.0812, -6.3193, -6.3295, -6.3297, -4.9219, -6.1962, -7.1518, -6.1548, -6.1548, -4.4753, -4.1174, -6.1026, -4.3243, -3.0032, -7.1022, -6.0997, -1.8249, -7.2521, -6.5448, -7.2872, -1.8694, -5.2641, -4.442, -4.0632, -5.0826, -4.7206, -2.7631, -3.9064, -4.3611, -4.7669, -4.6862, -4.2733, -4.9762, -5.081, -5.1217, -4.968, -5.0904, -5.0907, -4.8996, -5.5654, -5.6016, -5.6018, -4.1222, -5.1923, -6.0152, -5.611, -4.7261, -4.763, -6.1467, -6.3133, -5.8352, -4.6725, -6.2226, -5.0097, -6.5081, -4.8548, -5.2187, -6.0427, -6.4445, -6.4449, -5.1623, -5.6594, -5.895, -6.6354, -6.1068, -5.3731, -6.5737, -4.8609, -5.9232, -3.6634, -4.2911, -4.672, -3.019, -5.854, -3.7778, -3.7974, -3.7985, -2.3866, -4.5605, -4.4902, -4.7776, -4.5403, -4.5597, -3.8152, -3.0156, -3.1941, -5.0291, -5.0103, -4.4148, -4.2222, -4.6101, -4.9336, -4.9587, -4.5323, -5.0165, -4.7402, -6.3136, -6.3147, -6.315, -6.3154, -5.3939, -5.997, -5.3098, -5.8567, -6.4574, -4.053, -5.4893, -5.635, -6.5234, -3.9649, -5.4336, -5.2922, -6.1168, -7.3184, -7.0771, -7.1462, -4.7179, -5.7738, -5.78, -7.1416, -7.3185, -3.8793, -3.8803, -3.8816, -3.8829, -3.8851, -4.7144, -3.8228, -4.6936, -2.9684, -2.6446, -3.9742, -4.6067, -3.6994, -3.5572, -5.3837, -5.4451, -4.9143, -4.2518, -5.1669, -4.764, -5.0793, -4.9255, -4.9715, -4.9775, -3.9136, -4.8004, -4.7522, -4.9599, -4.8026, -5.4232, -5.6899, -5.8287, -5.8287, -5.8287, -5.5462, -5.8131, -6.1596, -4.5731, -4.5763, -6.6638, -4.5693, -5.2902, -6.87, -4.9706, -5.2091, -6.0413, -5.5759, -6.2321, -6.8255, -6.3944, -7.0703, -4.5721, -5.4314, -5.1639, -6.4753, -5.7635, -4.9897, -5.575, -5.5752, -4.9856, -4.9856, -4.9856, -3.8197, -3.6995, -4.9856, -3.9378, -4.8082, -4.5743, -4.5786, -4.5834, -4.5485, -4.5693, -4.6496, -4.6496, -3.9825, -4.6493, -2.7276, -4.2098, -3.9929, -3.9939, -3.6575, -4.048, -4.5966, -4.5491, -3.8121, -4.2355, -4.5668, -4.6638, -4.5951, -4.4981, -4.4108, -5.0937, -4.1643, -5.5285, -5.7819, -5.1432, -5.1432, -5.1432, -6.1345, -3.2298, -6.1255, -6.2711, -5.3911, -4.6928, -6.5307, -6.531, -6.5311, -6.2399, -6.642, -6.5313, -5.0187, -6.2931, -4.7616, -3.6807, -5.4011, -5.096, -6.2637, -4.4917, -4.9889, -4.9123, -6.4267, -4.7682, -3.9199, -3.9998, -3.7724, -3.0304, -3.9115, -5.186, -5.3167, -4.5966, -3.9584, -4.1273, -5.3413, -4.7366, -3.4768, -4.445, -4.029, -4.3894, -4.9593, -3.7669, -4.1849, -4.4716, -4.4287, -4.6591, -4.8863, -4.9256, -4.9436], \"Category\": [\"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Default\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic1\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic2\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic3\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic4\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\", \"Topic5\"], \"Term\": [\"lack\", \"assurance\", \"tablet\", \"repackaged\", \"penicillin\", \"mg\", \"presence\", \"potency\", \"recall\", \"within_expiry\", \"product\", \"specification_result\", \"pedigree\", \"stability_data_do_not\", \"drug_package\", \"fda_inspection_identify_gmp\", \"violation_potentially_impact\", \"without_adequate_separation_which\", \"potential_for_cross_contamination\", \"also_cgmp_deviation\", \"form\", \"could_introduce\", \"market_without_an_approved\", \"particulate_matter\", \"adverse_reaction\", \"firm_receive_seven_report\", \"80mg/ml_10_ml_vial\", \"support_expiry_potential_loss\", \"compound_preservative_free_methylprednisolone\", \"skin_abscess_potentially_link\", \"without_adequate_separation_which\", \"potential_for_cross_contamination\", \"could_introduce\", \"produce\", \"also_cgmp_deviation\", \"repackaged\", \"02/12/15\", \"distribute_between_01/05/12\", \"facility\", \"inspection_observation_associate\", \"support_expiry_recent\", \"not_manufacture_accord\", \"cgmp_deviation_pharmaceutical\", \"good_manufacturing_practices\", \"processing_controls\", \"guarantee\", \"distribute_by_this\", \"content_uniformity_failure\", \"assay_or\", \"store\", \"recall_because_they\", \"not_assure\", \"compound\", \"penicillin\", \"quality_control_process\", \"distribute\", \"lack\", \"customer\", \"ml\", \"laboratory\", \"assurance\", \"manufacturer\", \"cgmp_deviation\", \"cross_contamination\", \"glass_particulate\", \"syringe\", \"product\", \"lot\", \"drug\", \"stability_data_do_not\", \"use\", \"quality\", \"sterile\", \"drug_package\", \"support_expiry_potential_loss\", \"recall\", \"fda_inspection_identify_gmp\", \"violation_potentially_impact\", \"affect\", \"withdraw_from\", \"fda_inspection_finding_result\", \"concern_regard_quality_control\", \"all_sterile_human\", \"active_ingredient_that\", \"for_safety_reason\", \"labeling:label_mixup\", \"observe_during\", \"compound_by\", \"leakage\", \"quality_control_procedure\", \"6_month\", \"potential_risk\", \"remove_from\", \"pharmacy_that\", \"neck\", \"present\", \"not_expire_due\", \"u.s._market\", \"05/21/2012\", \"fda_environmental_sampling\", \"process\", \"contain_undeclared_sibutramine\", \"subject\", \"inspection\", \"broken\", \"number\", \"control\", \"select_sterile\", \"certain\", \"within_expiry\", \"firm\", \"vial\", \"recall\", \"glass\", \"quality\", \"fda_inspection_identify_gmp\", \"violation_potentially_impact\", \"product\", \"recent_fda_inspection\", \"initiate\", \"concern_associate\", \"sterile\", \"subpotent_drug\", \"lot\", \"assurance\", \"lack\", \"potential\", \"quality_control_procedure_that\", \"drug\", \"presence\", \"cross_contamination\", \"manufacture\", \"result\", \"failed_dissolution_specification\", \"know_impurity\", \"mislabeled_as_one\", \"contain_62%_ethyl_alcohol\", \"ethyl_alcohol_content\", \"label_state\", \"20%\", \"fail_impurity/degradation_specification\", \"lot_and/or_expiration\", \"lozenge\", \"exp_6/26/2014\", \"0.125\", \"may_have_potentially\", \"fda_inspectional_finding_result\", \"overly_thick_overly_soft\", \"ascorbic_acid\", \"follow_drug\", \"overwrap\", \"impact\", \"objectionable_condition_observe_during\", \"by_c._difficile_discover\", \"hyoscyamine_sulfate_sl\", \"00904052260_pedigree\", \"mislabeled_as\", \"iv_bag_leak\", \"puncture_through\", \"quetiapine_fumarate\", \"possible_microbial_contamination\", \"adverse_reaction\", \"firm_receive_seven_report\", \"80mg/ml_10_ml_vial\", \"compound_preservative_free_methylprednisolone\", \"skin_abscess_potentially_link\", \"defective_container\", \"form\", \"500\", \"mg\", \"tablet\", \"labeling_label_mixup\", \"10\", \"mg_ndc\", \"pedigree\", \"documentation\", \"rubber\", \"products\", \"bottle\", \"2\", \"microbial_contamination\", \"hcl\", \"non_sterile\", \"ndc\", \"potentially_mislabeled_as\", \"product\", \"contain\", \"specification\", \"result\", \"assurance\", \"panel\", \"active_sunscreen_ingredient\", \"drug/disease_claim_make_them\", \"labeling_that_bear\", \"mark_as_dietary_supplement\", \"salicylic_acid\", \"fda_sampling_confirm\", \"failed_lozenge_specifications\", \"connection\", \"martin_avenue_pharmacy_inc.\", \"not_elsewhere_classify\", \"conduct\", \"strength\", \"an_analogue\", \"package_insert\", \"instead\", \"have_an\", \"total_impurity\", \"less_than\", \"36_month\", \"room_temperature\", \"male_erectile\", \"voluntary\", \"potentially_contaminate\", \"correct\", \"sildenafil\", \"unapproved_drug\", \"stability\", \"approve_nda/anda\", \"market_without\", \"aseptic_practice\", \"fda_inspection_reveal_poor\", \"condition_potentially\", \"nda/anda\", \"market_without_an_approved\", \"their_compound\", \"label\", \"fda_inspection\", \"certain_quality_control_procedure\", \"that_present\", \"risk\", \"observation_associate\", \"compound_sterile_preparation\", \"produce_sterile\", \"particulate_matter_api_contaminate\", \"specification\", \"injectable_drug\", \"product\", \"use\", \"violation_potentially_impact\", \"fda_inspection_identify_gmp\", \"recall\", \"presence\", \"recent_fda_inspection\", \"initiate\", \"assurance\", \"tablet\", \"within_expiry\", \"find\", \"pedigree\", \"penicillin\", \"lack\", \"18_month_stability_time\", \"fail_impurities/degradation_specification\", \"stability_datum_do_not\", \"particulate_matter_confirm_customer\", \"blister_cavity_may\", \"/capsule\", \"contain_more_than_one\", \"contain_foreign_substance\", \"potency\", \"visible_particulate_matter\", \"interval\", \"related\", \"foreign_tablets/capsule\", \"medical_inc.\", \"particulate_matter_b._braun\", \"several_injectable\", \"float\", \"plastic\", \"reserve_sample_unit\", \"obtain\", \"black\", \"fail_impurities/degradation_specifications_out\", \"specification_result\", \"support_expiry\", \"foreign_substance\", \"identify_during\", \"impurity\", \"and/or_exp\", \"point\", \"illegible\", \"identify_as\", \"drug_package\", \"support_expiry_potential_loss\", \"particulate_matter\", \"presence\", \"stability_data_do_not\", \"superpotent_drug\", \"stability_time_point\", \"test\", \"find\", \"syringe\", \"temperature_abuse\", \"date\", \"tablet\", \"subpotent_drug\", \"mg\", \"specification\", \"labeling_incorrect_or_missing\", \"product\", \"recall\", \"pedigree\", \"lot\", \"mg_ndc\", \"microbial_contamination\", \"capsule\", \"potentially_mislabeled_as\"], \"loglift\": [30.0, 29.0, 28.0, 27.0, 26.0, 25.0, 24.0, 23.0, 22.0, 21.0, 20.0, 19.0, 18.0, 17.0, 16.0, 15.0, 14.0, 13.0, 12.0, 11.0, 10.0, 9.0, 8.0, 7.0, 6.0, 5.0, 4.0, 3.0, 2.0, 1.0, 1.2228, 1.2225, 1.2216, 1.2149, 1.2131, 1.208, 1.2055, 1.2051, 1.1936, 1.1915, 1.1893, 1.1762, 1.1755, 1.1623, 1.1435, 1.1338, 1.1216, 1.1, 1.1, 1.0997, 1.0912, 1.071, 1.0505, 1.0491, 1.0114, 1.0095, 0.9835, 0.9749, 0.957, 0.9152, 0.9017, 0.9151, 0.8859, 0.795, 0.8698, 0.6659, 0.0975, 0.3711, 0.4446, 0.5699, 0.4934, 0.3078, 0.3755, 0.4371, 0.4584, -1.0709, -0.4937, -0.4946, 1.3523, 1.3439, 1.3416, 1.3411, 1.3396, 1.3316, 1.3288, 1.3277, 1.3263, 1.3196, 1.3127, 1.3106, 1.3065, 1.3063, 1.305, 1.3034, 1.3005, 1.2964, 1.2954, 1.2925, 1.2912, 1.291, 1.2834, 1.2737, 1.2613, 1.2553, 1.2434, 1.2428, 1.235, 1.2284, 1.2253, 1.165, 1.1109, 1.0864, 0.8781, 1.2012, 0.8033, 0.7993, 0.7976, 0.474, 0.9019, 0.8208, 0.8971, 0.8114, 0.8043, 0.4623, -0.2445, -0.3857, 0.9412, 0.8951, 0.3909, 0.0639, 0.2481, 0.7176, 0.2768, 1.5419, 1.5345, 1.5207, 1.5143, 1.5139, 1.5139, 1.5137, 1.5099, 1.5095, 1.5055, 1.4889, 1.4883, 1.4796, 1.4768, 1.4696, 1.4688, 1.4658, 1.4561, 1.4548, 1.4505, 1.4473, 1.4459, 1.4412, 1.4293, 1.4217, 1.4197, 1.4184, 1.4138, 1.4111, 1.4108, 1.4101, 1.4098, 1.4087, 1.4017, 1.3828, 1.3528, 1.2015, 1.1577, 1.2539, 1.3155, 1.1765, 1.12, 1.3851, 1.3582, 1.1955, 0.9605, 1.2247, 1.0043, 1.1709, 1.0445, 1.0096, 0.9278, -1.0531, 0.485, 0.2198, 0.2756, -2.0315, 1.9498, 1.9377, 1.9275, 1.9275, 1.9275, 1.903, 1.8789, 1.8643, 1.8513, 1.8473, 1.8281, 1.8082, 1.7944, 1.7887, 1.7881, 1.7784, 1.7746, 1.7717, 1.7673, 1.76, 1.7454, 1.733, 1.7152, 1.6801, 1.6752, 1.663, 1.6488, 1.6334, 1.615, 1.6148, 1.5823, 1.5823, 1.5822, 1.5038, 1.4904, 1.5822, 1.4495, 1.5562, 1.506, 1.5014, 1.4945, 1.4693, 1.4448, 1.4227, 1.4227, 0.9895, 1.3248, 0.1329, 0.9698, 0.6032, 0.6028, 0.2396, 0.2381, 0.8658, 0.7619, -1.041, -0.4333, 0.2616, 0.5114, 0.0822, -0.4458, -1.6024, 2.1808, 2.1702, 2.16, 2.1177, 2.102, 2.102, 2.102, 2.0837, 2.0635, 2.0578, 2.0419, 2.0384, 2.0183, 1.9868, 1.9863, 1.9861, 1.9793, 1.9534, 1.9361, 1.9308, 1.9201, 1.9194, 1.8886, 1.8768, 1.8764, 1.8459, 1.82, 1.8042, 1.8006, 1.7661, 1.7086, 1.5982, 1.5803, 1.5023, 1.2557, 1.4253, 1.69, 1.7163, 1.4799, 1.2168, 1.2592, 1.6957, 1.2848, 0.3255, 0.919, 0.1409, 0.5826, 1.2935, -0.9064, -0.2877, 0.2057, -0.1512, 0.2168, 0.882, 0.9384, 0.9616], \"Freq\": [3341.0, 3468.0, 1236.0, 550.0, 963.0, 856.0, 762.0, 278.0, 1124.0, 443.0, 3172.0, 211.0, 515.0, 266.0, 222.0, 558.0, 559.0, 263.0, 263.0, 280.0, 303.0, 263.0, 308.0, 283.0, 279.0, 279.0, 278.0, 209.0, 278.0, 278.0, 260.5589289198424, 260.54461025177943, 260.444374144943, 84.44455340609811, 275.43556169226844, 536.5914783756252, 260.4823727748921, 258.7780181502134, 265.5890070739592, 36.94297519909198, 36.90797668185934, 29.08728803738237, 28.79244775999284, 28.788276438962306, 117.6481703331065, 32.900179570424086, 12.651991638790744, 34.28842909855028, 34.28842909855028, 183.88546482636178, 263.03193440989355, 36.12619240982652, 213.85985471752838, 801.4551366423624, 13.29515952704931, 36.23200659576335, 2603.7257415328345, 11.444927411878805, 23.215714728929186, 11.049962066670794, 2490.494742459, 83.56000871675941, 190.11705777769416, 277.6615368003535, 100.18533280909732, 143.88076255027556, 1018.988539867886, 324.80847857295413, 206.13904767618013, 137.37041181950946, 148.9253706828882, 225.05084568429226, 111.43498116105546, 100.35147453917918, 96.34176803361463, 112.34947718729933, 99.41217027070968, 99.38072479813431, 104.13468333530192, 53.50872257565713, 51.60839761566225, 51.59802455459089, 226.5736950664469, 77.70887254735067, 34.12545687470943, 51.123071704965035, 123.86061291782904, 119.37824037181062, 29.920778188414154, 25.330344252363158, 40.854376895269176, 130.67416523406274, 27.734589922521295, 93.27542041602057, 20.846588318208855, 108.90229048228018, 75.68294954616134, 33.20057079199178, 22.21445384831209, 22.20679353986991, 80.07547054681507, 48.70789389650919, 38.48450610424537, 18.354017261647527, 31.140047923424806, 64.85334611860351, 19.52191022181354, 108.23579333125872, 37.41266936385617, 358.4852856698105, 191.35762580013358, 130.74846240256838, 682.8172860526737, 40.096012027075595, 319.72197465906675, 313.5154316728616, 313.17645890890776, 1285.2240523340042, 146.16257352098884, 156.81724966747615, 117.63862595746018, 149.1507836845708, 146.29155892180657, 307.9929234073701, 685.1742646994878, 573.1525522507052, 91.47994640261491, 93.22347215670617, 169.10114955137507, 205.002922893361, 139.08962957288549, 100.65541644160491, 98.15714858726405, 125.74058029538868, 77.47892476220939, 102.13762266636188, 21.17729170930852, 21.154203045367613, 21.147150109127487, 21.13823545162884, 53.1245277771221, 29.065858630543435, 57.78290561662953, 33.44097372664942, 18.340333293288666, 203.0696822321227, 48.29231335258876, 41.74072801437443, 17.168755980388614, 221.7585175983484, 51.0558529096793, 58.81220415376055, 25.782381273772558, 7.753485340024253, 9.869022648585048, 9.20988183987986, 104.44343312677212, 36.332208167846474, 36.10767742787927, 9.253099121011868, 7.752838083677037, 241.58830886928348, 241.33398727678824, 241.02482247394252, 240.7285604788883, 240.1988417115746, 104.8051500110375, 255.62339754887702, 107.0102480159288, 600.7023043643405, 830.4127395486495, 219.711194377885, 116.73038176522446, 289.19431803304485, 333.39335467234724, 53.667629399257535, 50.47346572681389, 85.81628461036367, 166.46345127848423, 66.6621529916432, 99.7376541368865, 72.76263837114512, 84.86584721557816, 81.04545576976277, 80.56586207449786, 233.44536984084547, 96.17286159383082, 100.92233192723498, 81.99419492754885, 95.96089744457278, 33.416872866454206, 25.59372992387195, 22.276294043183224, 22.276294043183224, 22.276294043183224, 29.54957019482001, 22.62566465319114, 16.00054096729414, 78.18525417468072, 77.93608958890253, 9.664442267267537, 78.48254617325576, 38.16954151286879, 7.863757334637346, 52.54311958917745, 41.39413275005164, 18.00904314035649, 28.68302590545885, 14.882098193866081, 8.221207052978986, 12.651707734512435, 6.436147106514358, 78.26421163572196, 33.14175201169927, 43.30879548902516, 11.668769782207011, 23.778078555348994, 51.546860181367684, 28.70899111937868, 28.704295682664323, 51.760956193627344, 51.759167146799854, 51.75978454356337, 166.08695050533868, 187.30806362844453, 51.75877632520259, 147.58163212264688, 61.80666746832397, 78.09520511492555, 77.76292693156653, 77.39078246530488, 80.13374349935495, 78.48369853565708, 72.42856937530046, 72.42856937530046, 141.13861348916228, 72.4488690600866, 495.0341028279726, 112.43734494729735, 139.6778963293981, 139.53662786633348, 195.32782381388336, 132.19316670191222, 76.36892255607559, 80.09091937550424, 167.36114555527172, 109.5832964735513, 78.67983504298152, 71.41211612437702, 76.48700951959793, 84.27768641309822, 91.96485405956454, 36.9838157232402, 93.67514644683297, 23.943127066900956, 18.583257594555203, 35.19722197106377, 35.19722197106377, 35.19722197106377, 13.061111983679021, 238.50781823061575, 13.180270193525574, 11.394145984949299, 27.46983944638794, 55.22250488578016, 8.788553222343541, 8.785798599367958, 8.785065148553699, 11.755430466744878, 7.862999270776711, 8.783459471080647, 39.862025556893876, 11.146125856347071, 51.55117049480269, 151.9414475560512, 27.19535678684518, 36.89932003522952, 11.47796697322245, 67.52634194627944, 41.07039553496904, 44.339909825429544, 9.751714618951226, 51.209523537586676, 119.61156968861954, 110.42721188576016, 138.6319573972982, 291.1363078664377, 120.61818682318555, 33.721201175403124, 29.589834695716895, 60.80202687880745, 115.09898942290131, 97.21211916300909, 28.871664778372793, 52.853746444556684, 186.30856738288028, 70.75264843130125, 107.2479069320606, 74.79767886111274, 42.303608431305186, 139.38766528209695, 91.77257377327199, 68.89644971288577, 71.91068864746724, 57.11599052539707, 45.507328489751075, 43.75575610064298, 42.97173337690545], \"Total\": [3341.0, 3468.0, 1236.0, 550.0, 963.0, 856.0, 762.0, 278.0, 1124.0, 443.0, 3172.0, 211.0, 515.0, 266.0, 222.0, 558.0, 559.0, 263.0, 263.0, 280.0, 303.0, 263.0, 308.0, 283.0, 279.0, 279.0, 278.0, 209.0, 278.0, 278.0, 263.23006585389817, 263.3057305486299, 263.4508249244875, 85.99222183887287, 280.98010657151696, 550.1991519182467, 267.7532036162128, 266.10757312906145, 276.26774306926, 38.51070474055526, 38.556535204008775, 30.78843608318151, 30.49597460308551, 30.896724330204222, 128.66152144713985, 36.33088028512648, 14.143229506411176, 39.16802217667908, 39.16802217667908, 210.11506057900104, 303.10103164291024, 42.48046947413476, 256.68902819065386, 963.2795898610888, 16.594200595935934, 45.305820975253965, 3341.681687138222, 14.814807853547174, 30.594352920111692, 15.184789212177746, 3468.7817787284807, 114.82904556865903, 269.0172873913113, 430.288641753436, 144.0609969946639, 253.69716395526433, 3172.0797301528055, 769.0445728053156, 453.5005803076524, 266.6245888796734, 312.02243671816933, 567.6520625520761, 262.69826480031105, 222.42138210817313, 209.0455675357997, 1124.9978500096297, 558.914583138879, 559.2586535918517, 106.7873623628602, 55.33176723180227, 53.49139708256522, 53.50505503918185, 235.3038312869724, 81.35465902645896, 35.82660213740922, 53.72771176404467, 130.3511413774098, 126.48483155063154, 31.92066739341124, 27.08054444642188, 43.85847233465365, 140.29989377432506, 29.816660882477585, 100.44499092531404, 22.51232568926109, 118.08659167478739, 82.15448765429353, 36.14377787712176, 24.21508235311908, 24.211440261951445, 87.97188334853648, 54.02940545939961, 43.22225062509316, 20.738639374689107, 35.60614174921854, 74.20101844011225, 22.509037017389616, 125.62423625409915, 43.5584095723696, 443.3101027524799, 249.79831730668232, 174.90599212104183, 1124.9978500096297, 47.82293990109055, 567.6520625520761, 558.914583138879, 559.2586535918517, 3172.0797301528055, 235.153528486361, 273.59752853009456, 190.1677184437835, 262.69826480031105, 259.47583519807495, 769.0445728053156, 3468.7817787284807, 3341.681687138222, 141.50031378240317, 151.01094964412502, 453.5005803076524, 762.4798998515321, 430.288641753436, 194.70219032124695, 295.052456905687, 127.53745566389526, 79.16946219692177, 105.82370536019823, 22.081483347581482, 22.06575452219629, 22.06027375104156, 22.053998499550485, 55.640782804527696, 30.454505822808233, 60.782486478440966, 35.765377034749996, 19.627745159827043, 219.2302166072518, 52.28173391784026, 45.511416878136494, 18.73540465448449, 242.7144307543287, 56.42678867233115, 65.08583389909045, 28.655291399525098, 8.645384596303916, 11.019007828055189, 10.332027640661348, 118.56362789717376, 41.56039881366818, 41.386580549310494, 10.619655090277389, 8.938707269113122, 279.28144121053464, 279.0726769966334, 278.93082844852637, 278.6658484289426, 278.3689447570695, 122.30950735988048, 303.9916520165433, 131.13560273424102, 856.3885047612007, 1236.9153928157652, 297.2294493843097, 148.4906611196371, 422.7361784478548, 515.6394263128445, 63.680129934280124, 61.51891476125102, 123.08403286197351, 302.00490029253126, 92.86221207595663, 173.19050728285868, 106.9629364023793, 141.55080719217173, 139.98566177457053, 151.0177430252982, 3172.0797301528055, 280.7030197612076, 384.01326190793264, 295.052456905687, 3468.7817787284807, 34.80401381397105, 26.97854008193313, 23.724544166555084, 23.724544166555084, 23.724544166555084, 32.24968695760118, 25.29492659958601, 18.151900501081947, 89.86045255135829, 89.92537861615516, 11.367623761594542, 94.17353180469115, 46.436262724150176, 9.621421413202508, 64.32516698885708, 51.17242398200384, 22.346926016051256, 35.69726571560217, 18.602687709189365, 10.352123105336352, 16.164183514511414, 8.325706054886604, 103.05827791384736, 45.19980498446815, 59.36074894344073, 16.189870166377784, 33.461192476545015, 73.66791519802892, 41.79083416430862, 41.7900366377536, 77.84632555329182, 77.8475095459226, 77.85620134468624, 270.2036583129726, 308.8226685778514, 77.8574984648014, 253.47980527189887, 95.41851249114379, 126.771927068825, 126.81020846533288, 127.08212140960846, 134.93714247230002, 135.44202006077137, 127.78485173551687, 127.78485173551687, 384.01326190793264, 140.96879875255487, 3172.0797301528055, 312.02243671816933, 559.2586535918517, 558.914583138879, 1124.9978500096297, 762.4798998515321, 235.153528486361, 273.59752853009456, 3468.7817787284807, 1236.9153928157652, 443.3101027524799, 313.40345631767656, 515.6394263128445, 963.2795898610888, 3341.681687138222, 38.40590195632411, 98.31074198724048, 25.385408604767683, 20.55449310364728, 39.545565219066546, 39.545565219066546, 39.545565219066546, 14.94531528309747, 278.4998451470292, 15.477132995380455, 13.59505928706118, 32.8898667805211, 67.46365683756386, 11.07977806030104, 11.082032129104395, 11.08333703726481, 14.931611007856265, 10.250215101768482, 11.648981083012837, 53.15056482160766, 15.021643639510105, 69.5265914835337, 211.32051622338741, 38.27130371741877, 51.95115441068938, 16.66017597675863, 100.58023441967885, 62.15035700076865, 67.34033492535464, 15.33024698612654, 85.26656735698653, 222.42138210817313, 209.0455675357997, 283.7362209184349, 762.4798998515321, 266.6245888796734, 57.20182698104127, 48.89104296321429, 127.25953496153589, 313.40345631767656, 253.69716395526433, 48.697796626287584, 134.4538156242282, 1236.9153928157652, 259.47583519807495, 856.3885047612007, 384.01326190793264, 106.6831334624083, 3172.0797301528055, 1124.9978500096297, 515.6394263128445, 769.0445728053156, 422.7361784478548, 173.19050728285868, 157.37998645137267, 151.0177430252982]}, \"plot.opts\": {\"xlab\": \"PC1\", \"ylab\": \"PC2\"}, \"topic.order\": [3, 2, 4, 5, 1], \"R\": 30, \"lambda.step\": 0.01};\n", | |
| "\n", | |
| "function LDAvis_load_lib(url, callback){\n", | |
| " var s = document.createElement('script');\n", | |
| " s.src = url;\n", | |
| " s.async = true;\n", | |
| " s.onreadystatechange = s.onload = callback;\n", | |
| " s.onerror = function(){console.warn(\"failed to load library \" + url);};\n", | |
| " document.getElementsByTagName(\"head\")[0].appendChild(s);\n", | |
| "}\n", | |
| "\n", | |
| "if(typeof(LDAvis) !== \"undefined\"){\n", | |
| " // already loaded: just create the visualization\n", | |
| " !function(LDAvis){\n", | |
| " new LDAvis(\"#\" + \"ldavis_el1013626482925773521468873287\", ldavis_el1013626482925773521468873287_data);\n", | |
| " }(LDAvis);\n", | |
| "}else if(typeof define === \"function\" && define.amd){\n", | |
| " // require.js is available: use it to load d3/LDAvis\n", | |
| " require.config({paths: {d3: \"https://cdnjs.cloudflare.com/ajax/libs/d3/3.5.5/d3.min\"}});\n", | |
| " require([\"d3\"], function(d3){\n", | |
| " window.d3 = d3;\n", | |
| " LDAvis_load_lib(\"https://cdn.rawgit.com/bmabey/pyLDAvis/files/ldavis.v1.0.0.js\", function(){\n", | |
| " new LDAvis(\"#\" + \"ldavis_el1013626482925773521468873287\", ldavis_el1013626482925773521468873287_data);\n", | |
| " });\n", | |
| " });\n", | |
| "}else{\n", | |
| " // require.js not available: dynamically load d3 & LDAvis\n", | |
| " LDAvis_load_lib(\"https://cdnjs.cloudflare.com/ajax/libs/d3/3.5.5/d3.min.js\", function(){\n", | |
| " LDAvis_load_lib(\"https://cdn.rawgit.com/bmabey/pyLDAvis/files/ldavis.v1.0.0.js\", function(){\n", | |
| " new LDAvis(\"#\" + \"ldavis_el1013626482925773521468873287\", ldavis_el1013626482925773521468873287_data);\n", | |
| " })\n", | |
| " });\n", | |
| "}\n", | |
| "</script>" | |
| ], | |
| "text/plain": [ | |
| "<IPython.core.display.HTML object>" | |
| ] | |
| }, | |
| "execution_count": 52, | |
| "metadata": {}, | |
| "output_type": "execute_result" | |
| } | |
| ], | |
| "source": [ | |
| "pyLDAvis.display(LDAvis_prepared)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "### Describing text with LDA\n", | |
| "Beyond data exploration, one of the key uses for an LDA model is providing a compact, quantitative description of natural language text. Once an LDA model has been trained, it can be used to represent free text as a mixture of the topics the model learned from the original corpus. This mixture can be interpreted as a probability distribution across the topics, so the LDA representation of a paragraph of text might look like 50% _Topic A_, 20% _Topic B_, 20% _Topic C_, and 10% _Topic D_.\n", | |
| "\n", | |
| "To use an LDA model to generate a vector representation of new text, you'll need to apply any text preprocessing steps you used on the model's training corpus to the new text, too. For our model, the preprocessing steps we used include:\n", | |
| "1. Using spaCy to remove punctuation and lemmatize the text\n", | |
| "1. Applying our first-order phrase model to join word pairs\n", | |
| "1. Applying our second-order phrase model to join longer phrases\n", | |
| "1. Removing stopwords\n", | |
| "1. Creating a bag-of-words representation\n", | |
| "\n", | |
| "Once you've applied these preprocessing steps to the new text, it's ready to pass directly to the model to create an LDA representation. The `lda_description(...)` function will perform all these steps for us, including printing the resulting topical description of the input text." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 53, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "def get_sample_recall(recall_number):\n", | |
| " \"\"\"\n", | |
| " retrieve a particular review index\n", | |
| " from the reviews file and return it\n", | |
| " \"\"\"\n", | |
| " \n", | |
| " return list(it.islice(line_review(recall_txt_filepath),\n", | |
| " recall_number, recall_number+1))[0]" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 54, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "def lda_description(recall_text, min_topic_freq=0.05):\n", | |
| " \"\"\"\n", | |
| " accept the original text of a review and (1) parse it with spaCy,\n", | |
| " (2) apply text pre-proccessing steps, (3) create a bag-of-words\n", | |
| " representation, (4) create an LDA representation, and\n", | |
| " (5) print a sorted list of the top topics in the LDA representation\n", | |
| " \"\"\"\n", | |
| " \n", | |
| " # parse the review text with spaCy\n", | |
| " parsed_reason = nlp(recall_text)\n", | |
| " \n", | |
| " # lemmatize the text and remove punctuation and whitespace\n", | |
| " unigram_reason = [token.lemma_ for token in parsed_reason\n", | |
| " if not punct_space(token)]\n", | |
| " \n", | |
| " # apply the first-order and secord-order phrase models\n", | |
| " bigram_reason = bigram_model[unigram_reason]\n", | |
| " trigram_reason = trigram_model[bigram_reason]\n", | |
| " \n", | |
| " # remove any remaining stopwords\n", | |
| " trigram_reason = [term for term in trigram_reason\n", | |
| " if not term in spacy.en.stop_words.STOP_WORDS]\n", | |
| " \n", | |
| " # create a bag-of-words representation\n", | |
| " reason_bow = trigram_dictionary.doc2bow(trigram_reason)\n", | |
| " \n", | |
| " # create an LDA representation\n", | |
| " reason_lda = lda[reason_bow]\n", | |
| " \n", | |
| " # sort with the most highly related topics first\n", | |
| " reason_lda = sorted(reason_lda, key=lambda topic_lda: -topic_lda[1])\n", | |
| " \n", | |
| " for topic_number, freq in reason_lda:\n", | |
| " if freq < min_topic_freq:\n", | |
| " break\n", | |
| " \n", | |
| " # print the most highly related topic names and frequencies\n", | |
| " print('{:2} {}'.format(topic_names[topic_number],\n", | |
| " round(freq, 2)))" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 55, | |
| "metadata": { | |
| "collapsed": false, | |
| "scrolled": true | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "Presence of Foreign Substance: Recall is being initiated due to the presence of a foreign particle identified from a customer complaint.\n", | |
| "\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "sample_reason = get_sample_recall(25)\n", | |
| "print(sample_reason)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 56, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "potency 0.79\n", | |
| "expiry 0.15\n" | |
| ] | |
| }, | |
| { | |
| "name": "stderr", | |
| "output_type": "stream", | |
| "text": [ | |
| "C:\\Users\\aashis_tiwari\\AppData\\Local\\Continuum\\Anaconda3\\envs\\tensorflow\\lib\\site-packages\\gensim\\models\\phrases.py:274: UserWarning: For a faster implementation, use the gensim.models.phrases.Phraser class\n", | |
| " warnings.warn(\"For a faster implementation, use the gensim.models.phrases.Phraser class\")\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "lda_description(sample_reason)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 57, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| " Lack of Assurance of Sterility: Franck's Lab Inc. initiated a recall of all Sterile Human Drugs distributed between 11/21/2011 and 05/21/2012. FDA environmental sampling revealed the presence of microorganisms and fungal growth in the clean room where sterile products were prepared. \n", | |
| "\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "sample_reason = get_sample_recall(100)\n", | |
| "print(sample_reason)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 58, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "expiry 0.95\n" | |
| ] | |
| }, | |
| { | |
| "name": "stderr", | |
| "output_type": "stream", | |
| "text": [ | |
| "C:\\Users\\aashis_tiwari\\AppData\\Local\\Continuum\\Anaconda3\\envs\\tensorflow\\lib\\site-packages\\gensim\\models\\phrases.py:274: UserWarning: For a faster implementation, use the gensim.models.phrases.Phraser class\n", | |
| " warnings.warn(\"For a faster implementation, use the gensim.models.phrases.Phraser class\")\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "lda_description(sample_reason)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "## Word Vector Embedding with Word2Vec" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Pop quiz! Can you complete this text snippet?\n", | |
| "\n", | |
| "<br><br>" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "<br><br><br>\n", | |
| "You just demonstrated the core machine learning concept behind word vector embedding models!\n", | |
| "<br><br><br>" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "The goal of *word vector embedding models*, or *word vector models* for short, is to learn dense, numerical vector representations for each term in a corpus vocabulary. If the model is successful, the vectors it learns about each term should encode some information about the *meaning* or *concept* the term represents, and the relationship between it and other terms in the vocabulary. Word vector models are also fully unsupervised &mdash they learn all of these meanings and relationships solely by analyzing the text of the corpus, without any advance knowledge provided.\n", | |
| "\n", | |
| "Perhaps the best-known word vector model is [word2vec](https://arxiv.org/pdf/1301.3781v3.pdf), originally proposed in 2013. The general idea of word2vec is, for a given *focus word*, to use the *context* of the word — i.e., the other words immediately before and after it — to provide hints about what the focus word might mean. To do this, word2vec uses a *sliding window* technique, where it considers snippets of text only a few tokens long at a time.\n", | |
| "\n", | |
| "At the start of the learning process, the model initializes random vectors for all terms in the corpus vocabulary. The model then slides the window across every snippet of text in the corpus, with each word taking turns as the focus word. Each time the model considers a new snippet, it tries to learn some information about the focus word based on the surrouding context, and it \"nudges\" the words' vector representations accordingly. One complete pass sliding the window across all of the corpus text is known as a training *epoch*. It's common to train a word2vec model for multiple passes/epochs over the corpus. Over time, the model rearranges the terms' vector representations such that terms that frequently appear in similar contexts have vector representations that are *close* to each other in vector space.\n", | |
| "\n", | |
| "For a deeper dive into word2vec's machine learning process, see [here](https://arxiv.org/pdf/1411.2738v4.pdf).\n", | |
| "\n", | |
| "Word2vec has a number of user-defined hyperparameters, including:\n", | |
| "- The dimensionality of the vectors. Typical choices include a few dozen to several hundred.\n", | |
| "- The width of the sliding window, in tokens. Five is a common default choice, but narrower and wider windows are possible.\n", | |
| "- The number of training epochs.\n", | |
| "\n", | |
| "For using word2vec in Python, [gensim](https://rare-technologies.com/deep-learning-with-word2vec-and-gensim/) comes to the rescue again! It offers a [highly-optimized](https://rare-technologies.com/word2vec-in-python-part-two-optimizing/), [parallelized](https://rare-technologies.com/parallelizing-word2vec-in-python/) implementation of the word2vec algorithm with its [Word2Vec](https://radimrehurek.com/gensim/models/word2vec.html) class." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 59, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "from gensim.models import Word2Vec\n", | |
| "\n", | |
| "trigram_sentences = LineSentence(trigram_sentences_filepath)\n", | |
| "word2vec_filepath = os.path.join(intermediate_directory, 'word2vec_model_all')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "We'll train our word2vec model using the normalized sentences with our phrase models applied. We'll use 100-dimensional vectors, and set up our training process to run for twelve epochs." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 110, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "12 training epochs so far.\n", | |
| "Wall time: 17.8 s\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "%%time\n", | |
| "\n", | |
| "# this is a bit time consuming - make the if statement True\n", | |
| "# if you want to train the word2vec model yourself.\n", | |
| "if 1 == 1:\n", | |
| "\n", | |
| " # initiate the model and perform the first epoch of training\n", | |
| " recall2vec = Word2Vec(trigram_sentences, size=100, window=5,\n", | |
| " min_count=20, sg=1, workers=1)\n", | |
| " \n", | |
| " recall2vec.save(word2vec_filepath)\n", | |
| "\n", | |
| " # perform another 11 epochs of training\n", | |
| " for i in range(1,12):\n", | |
| "\n", | |
| " recall2vec.train(trigram_sentences)\n", | |
| " recall2vec.save(word2vec_filepath)\n", | |
| " \n", | |
| "# load the finished model from disk\n", | |
| "recall2vec = Word2Vec.load(word2vec_filepath)\n", | |
| "recall2vec.init_sims()\n", | |
| "\n", | |
| "print('{} training epochs so far.'.format(recall2vec.train_count))" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 111, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "422 terms in the recall2vec vocabulary.\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "print('{:,} terms in the recall2vec vocabulary.'.format(len(recall2vec.wv.vocab)))" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Let's take a peek at the word vectors our model has learned. We'll create a pandas DataFrame with the terms as the row labels, and the 100 dimensions of the word vector model as the columns." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 62, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "data": { | |
| "text/html": [ | |
| "<div>\n", | |
| "<table border=\"1\" class=\"dataframe\">\n", | |
| " <thead>\n", | |
| " <tr style=\"text-align: right;\">\n", | |
| " <th></th>\n", | |
| " <th>0</th>\n", | |
| " <th>1</th>\n", | |
| " <th>2</th>\n", | |
| " <th>3</th>\n", | |
| " <th>4</th>\n", | |
| " <th>5</th>\n", | |
| " <th>6</th>\n", | |
| " <th>7</th>\n", | |
| " <th>8</th>\n", | |
| " <th>9</th>\n", | |
| " <th>...</th>\n", | |
| " <th>90</th>\n", | |
| " <th>91</th>\n", | |
| " <th>92</th>\n", | |
| " <th>93</th>\n", | |
| " <th>94</th>\n", | |
| " <th>95</th>\n", | |
| " <th>96</th>\n", | |
| " <th>97</th>\n", | |
| " <th>98</th>\n", | |
| " <th>99</th>\n", | |
| " </tr>\n", | |
| " </thead>\n", | |
| " <tbody>\n", | |
| " <tr>\n", | |
| " <th>of</th>\n", | |
| " <td>0.250408</td>\n", | |
| " <td>-0.426319</td>\n", | |
| " <td>-0.465205</td>\n", | |
| " <td>-0.204381</td>\n", | |
| " <td>-0.392508</td>\n", | |
| " <td>2.501663e-01</td>\n", | |
| " <td>-0.158044</td>\n", | |
| " <td>0.304148</td>\n", | |
| " <td>0.218495</td>\n", | |
| " <td>0.383973</td>\n", | |
| " <td>...</td>\n", | |
| " <td>-0.232480</td>\n", | |
| " <td>-0.333301</td>\n", | |
| " <td>0.318591</td>\n", | |
| " <td>0.029734</td>\n", | |
| " <td>-0.192704</td>\n", | |
| " <td>-0.228435</td>\n", | |
| " <td>-0.041155</td>\n", | |
| " <td>-0.158670</td>\n", | |
| " <td>0.077506</td>\n", | |
| " <td>-0.346080</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>be</th>\n", | |
| " <td>0.349246</td>\n", | |
| " <td>-0.090556</td>\n", | |
| " <td>-0.084044</td>\n", | |
| " <td>-0.466857</td>\n", | |
| " <td>-0.324749</td>\n", | |
| " <td>-5.094894e-02</td>\n", | |
| " <td>-0.038947</td>\n", | |
| " <td>0.237301</td>\n", | |
| " <td>0.152482</td>\n", | |
| " <td>0.056053</td>\n", | |
| " <td>...</td>\n", | |
| " <td>-0.071386</td>\n", | |
| " <td>0.110380</td>\n", | |
| " <td>-0.113990</td>\n", | |
| " <td>0.206021</td>\n", | |
| " <td>0.123648</td>\n", | |
| " <td>-0.001053</td>\n", | |
| " <td>-0.013274</td>\n", | |
| " <td>0.230379</td>\n", | |
| " <td>-0.141617</td>\n", | |
| " <td>-0.203462</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>the</th>\n", | |
| " <td>0.067424</td>\n", | |
| " <td>0.080021</td>\n", | |
| " <td>0.033621</td>\n", | |
| " <td>0.182714</td>\n", | |
| " <td>-0.021266</td>\n", | |
| " <td>-5.362771e-01</td>\n", | |
| " <td>0.272588</td>\n", | |
| " <td>0.268369</td>\n", | |
| " <td>-0.154391</td>\n", | |
| " <td>0.496174</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.333724</td>\n", | |
| " <td>-0.015216</td>\n", | |
| " <td>0.251310</td>\n", | |
| " <td>0.469194</td>\n", | |
| " <td>0.128453</td>\n", | |
| " <td>0.241228</td>\n", | |
| " <td>-0.147339</td>\n", | |
| " <td>-0.153858</td>\n", | |
| " <td>0.165506</td>\n", | |
| " <td>-0.316384</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>sterility</th>\n", | |
| " <td>0.303488</td>\n", | |
| " <td>-0.228153</td>\n", | |
| " <td>-0.219394</td>\n", | |
| " <td>-0.302188</td>\n", | |
| " <td>-0.070613</td>\n", | |
| " <td>4.507700e-01</td>\n", | |
| " <td>-0.125470</td>\n", | |
| " <td>-0.174656</td>\n", | |
| " <td>0.334442</td>\n", | |
| " <td>0.810358</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.372313</td>\n", | |
| " <td>0.377984</td>\n", | |
| " <td>-0.309888</td>\n", | |
| " <td>-0.746572</td>\n", | |
| " <td>-0.342844</td>\n", | |
| " <td>-0.191247</td>\n", | |
| " <td>0.163880</td>\n", | |
| " <td>0.044130</td>\n", | |
| " <td>0.070255</td>\n", | |
| " <td>-1.068096</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>assurance</th>\n", | |
| " <td>0.240375</td>\n", | |
| " <td>-0.008034</td>\n", | |
| " <td>-0.594212</td>\n", | |
| " <td>0.349680</td>\n", | |
| " <td>-0.603484</td>\n", | |
| " <td>5.307044e-01</td>\n", | |
| " <td>0.029388</td>\n", | |
| " <td>0.626848</td>\n", | |
| " <td>0.348065</td>\n", | |
| " <td>0.077353</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.388538</td>\n", | |
| " <td>0.002734</td>\n", | |
| " <td>-0.239271</td>\n", | |
| " <td>-0.082077</td>\n", | |
| " <td>-0.047349</td>\n", | |
| " <td>-0.296976</td>\n", | |
| " <td>-0.091350</td>\n", | |
| " <td>0.045781</td>\n", | |
| " <td>0.393641</td>\n", | |
| " <td>-0.406683</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>product</th>\n", | |
| " <td>0.167806</td>\n", | |
| " <td>-0.021859</td>\n", | |
| " <td>0.185118</td>\n", | |
| " <td>-0.030679</td>\n", | |
| " <td>-0.188094</td>\n", | |
| " <td>1.409636e-01</td>\n", | |
| " <td>-0.239155</td>\n", | |
| " <td>0.544782</td>\n", | |
| " <td>0.352739</td>\n", | |
| " <td>0.016653</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.025098</td>\n", | |
| " <td>-0.186757</td>\n", | |
| " <td>-0.083697</td>\n", | |
| " <td>-0.093634</td>\n", | |
| " <td>-0.106546</td>\n", | |
| " <td>-0.198794</td>\n", | |
| " <td>0.290512</td>\n", | |
| " <td>0.423234</td>\n", | |
| " <td>-0.167009</td>\n", | |
| " <td>-0.739767</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>lack</th>\n", | |
| " <td>0.666243</td>\n", | |
| " <td>-0.235590</td>\n", | |
| " <td>-0.383695</td>\n", | |
| " <td>0.450461</td>\n", | |
| " <td>-0.533766</td>\n", | |
| " <td>6.104476e-01</td>\n", | |
| " <td>-0.394387</td>\n", | |
| " <td>0.135181</td>\n", | |
| " <td>-0.221998</td>\n", | |
| " <td>0.284320</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.096456</td>\n", | |
| " <td>0.144373</td>\n", | |
| " <td>0.413397</td>\n", | |
| " <td>-0.051289</td>\n", | |
| " <td>-0.165963</td>\n", | |
| " <td>-0.059666</td>\n", | |
| " <td>-0.216124</td>\n", | |
| " <td>-0.388248</td>\n", | |
| " <td>0.065185</td>\n", | |
| " <td>-0.543164</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>and</th>\n", | |
| " <td>0.013336</td>\n", | |
| " <td>0.060998</td>\n", | |
| " <td>-0.056626</td>\n", | |
| " <td>0.209782</td>\n", | |
| " <td>0.027815</td>\n", | |
| " <td>2.330013e-02</td>\n", | |
| " <td>-0.311109</td>\n", | |
| " <td>0.436705</td>\n", | |
| " <td>-0.339965</td>\n", | |
| " <td>0.205782</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.457187</td>\n", | |
| " <td>-0.042881</td>\n", | |
| " <td>0.268263</td>\n", | |
| " <td>-0.257201</td>\n", | |
| " <td>0.311483</td>\n", | |
| " <td>-0.102934</td>\n", | |
| " <td>0.405758</td>\n", | |
| " <td>0.062987</td>\n", | |
| " <td>-0.083985</td>\n", | |
| " <td>-0.631277</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>in</th>\n", | |
| " <td>0.106915</td>\n", | |
| " <td>0.433992</td>\n", | |
| " <td>-0.022223</td>\n", | |
| " <td>-0.229789</td>\n", | |
| " <td>-0.483802</td>\n", | |
| " <td>-2.567068e-01</td>\n", | |
| " <td>-0.353319</td>\n", | |
| " <td>-0.128223</td>\n", | |
| " <td>-0.179633</td>\n", | |
| " <td>0.151007</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.287440</td>\n", | |
| " <td>-0.491665</td>\n", | |
| " <td>0.040266</td>\n", | |
| " <td>0.345051</td>\n", | |
| " <td>0.493042</td>\n", | |
| " <td>0.187467</td>\n", | |
| " <td>0.664412</td>\n", | |
| " <td>0.068200</td>\n", | |
| " <td>-0.363377</td>\n", | |
| " <td>-0.660456</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>to</th>\n", | |
| " <td>-0.055777</td>\n", | |
| " <td>-0.142672</td>\n", | |
| " <td>-0.052781</td>\n", | |
| " <td>-0.160961</td>\n", | |
| " <td>-0.114285</td>\n", | |
| " <td>9.478991e-02</td>\n", | |
| " <td>0.197403</td>\n", | |
| " <td>0.107659</td>\n", | |
| " <td>0.009553</td>\n", | |
| " <td>0.110412</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.030555</td>\n", | |
| " <td>0.313960</td>\n", | |
| " <td>0.226491</td>\n", | |
| " <td>0.116636</td>\n", | |
| " <td>0.144624</td>\n", | |
| " <td>0.104488</td>\n", | |
| " <td>0.230591</td>\n", | |
| " <td>-0.263950</td>\n", | |
| " <td>-0.015821</td>\n", | |
| " <td>-0.146170</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>with</th>\n", | |
| " <td>0.139916</td>\n", | |
| " <td>0.056741</td>\n", | |
| " <td>0.019701</td>\n", | |
| " <td>-0.038652</td>\n", | |
| " <td>-0.131373</td>\n", | |
| " <td>3.501720e-02</td>\n", | |
| " <td>-0.514761</td>\n", | |
| " <td>0.129861</td>\n", | |
| " <td>0.129339</td>\n", | |
| " <td>0.307071</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.361517</td>\n", | |
| " <td>0.152685</td>\n", | |
| " <td>-0.730674</td>\n", | |
| " <td>0.113176</td>\n", | |
| " <td>0.702688</td>\n", | |
| " <td>0.153057</td>\n", | |
| " <td>0.254042</td>\n", | |
| " <td>-0.105156</td>\n", | |
| " <td>-0.114450</td>\n", | |
| " <td>0.147127</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>a</th>\n", | |
| " <td>0.515118</td>\n", | |
| " <td>0.419556</td>\n", | |
| " <td>0.291573</td>\n", | |
| " <td>-0.048714</td>\n", | |
| " <td>-0.854946</td>\n", | |
| " <td>2.778567e-01</td>\n", | |
| " <td>0.081357</td>\n", | |
| " <td>0.085357</td>\n", | |
| " <td>0.118027</td>\n", | |
| " <td>0.006842</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.433700</td>\n", | |
| " <td>0.406811</td>\n", | |
| " <td>-0.138931</td>\n", | |
| " <td>-0.100497</td>\n", | |
| " <td>0.305594</td>\n", | |
| " <td>-0.039583</td>\n", | |
| " <td>0.217508</td>\n", | |
| " <td>0.132062</td>\n", | |
| " <td>-0.141048</td>\n", | |
| " <td>0.212403</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>penicillin</th>\n", | |
| " <td>0.331125</td>\n", | |
| " <td>0.195782</td>\n", | |
| " <td>-0.216822</td>\n", | |
| " <td>-0.540945</td>\n", | |
| " <td>-0.070391</td>\n", | |
| " <td>-1.685262e-01</td>\n", | |
| " <td>0.119359</td>\n", | |
| " <td>0.104435</td>\n", | |
| " <td>-0.079008</td>\n", | |
| " <td>0.072145</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.097845</td>\n", | |
| " <td>0.443677</td>\n", | |
| " <td>-0.943778</td>\n", | |
| " <td>-0.739152</td>\n", | |
| " <td>0.490247</td>\n", | |
| " <td>-0.213078</td>\n", | |
| " <td>0.071543</td>\n", | |
| " <td>0.367104</td>\n", | |
| " <td>-0.180235</td>\n", | |
| " <td>-0.564265</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>all</th>\n", | |
| " <td>0.767341</td>\n", | |
| " <td>-0.377597</td>\n", | |
| " <td>-0.033244</td>\n", | |
| " <td>-0.079458</td>\n", | |
| " <td>-0.263991</td>\n", | |
| " <td>3.638779e-01</td>\n", | |
| " <td>-0.942502</td>\n", | |
| " <td>0.240766</td>\n", | |
| " <td>-0.142430</td>\n", | |
| " <td>0.114177</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.210978</td>\n", | |
| " <td>0.069528</td>\n", | |
| " <td>-0.270203</td>\n", | |
| " <td>-0.461119</td>\n", | |
| " <td>0.401482</td>\n", | |
| " <td>-0.016561</td>\n", | |
| " <td>-0.083103</td>\n", | |
| " <td>-0.058704</td>\n", | |
| " <td>0.028399</td>\n", | |
| " <td>-0.643742</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>recall</th>\n", | |
| " <td>-0.068916</td>\n", | |
| " <td>-0.640970</td>\n", | |
| " <td>0.074110</td>\n", | |
| " <td>-0.269826</td>\n", | |
| " <td>-0.126685</td>\n", | |
| " <td>1.882937e-01</td>\n", | |
| " <td>0.419718</td>\n", | |
| " <td>0.327580</td>\n", | |
| " <td>0.170750</td>\n", | |
| " <td>0.496003</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.107886</td>\n", | |
| " <td>0.658807</td>\n", | |
| " <td>0.094854</td>\n", | |
| " <td>0.475893</td>\n", | |
| " <td>0.054919</td>\n", | |
| " <td>0.181283</td>\n", | |
| " <td>0.224981</td>\n", | |
| " <td>0.670257</td>\n", | |
| " <td>0.409089</td>\n", | |
| " <td>-0.226304</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>tablet</th>\n", | |
| " <td>0.150178</td>\n", | |
| " <td>-0.334870</td>\n", | |
| " <td>-0.514988</td>\n", | |
| " <td>-0.383748</td>\n", | |
| " <td>-0.048423</td>\n", | |
| " <td>1.644191e-02</td>\n", | |
| " <td>0.362565</td>\n", | |
| " <td>-0.134630</td>\n", | |
| " <td>-0.053598</td>\n", | |
| " <td>-0.112347</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.610540</td>\n", | |
| " <td>0.415149</td>\n", | |
| " <td>0.225299</td>\n", | |
| " <td>0.186411</td>\n", | |
| " <td>0.109859</td>\n", | |
| " <td>0.216297</td>\n", | |
| " <td>0.002845</td>\n", | |
| " <td>0.259346</td>\n", | |
| " <td>-0.213895</td>\n", | |
| " <td>-0.033015</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>repackaged</th>\n", | |
| " <td>0.234697</td>\n", | |
| " <td>0.004605</td>\n", | |
| " <td>-0.236921</td>\n", | |
| " <td>-0.172754</td>\n", | |
| " <td>-0.094025</td>\n", | |
| " <td>-1.371086e-01</td>\n", | |
| " <td>-0.204735</td>\n", | |
| " <td>0.162600</td>\n", | |
| " <td>-0.507803</td>\n", | |
| " <td>0.215561</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.202148</td>\n", | |
| " <td>0.176992</td>\n", | |
| " <td>-0.694359</td>\n", | |
| " <td>-0.579576</td>\n", | |
| " <td>0.460001</td>\n", | |
| " <td>0.098583</td>\n", | |
| " <td>-0.406318</td>\n", | |
| " <td>0.367639</td>\n", | |
| " <td>-0.038518</td>\n", | |
| " <td>-0.390859</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>lot</th>\n", | |
| " <td>0.410613</td>\n", | |
| " <td>0.485917</td>\n", | |
| " <td>-0.436936</td>\n", | |
| " <td>-0.594825</td>\n", | |
| " <td>0.186103</td>\n", | |
| " <td>2.725144e-01</td>\n", | |
| " <td>-0.276863</td>\n", | |
| " <td>-0.037303</td>\n", | |
| " <td>0.059433</td>\n", | |
| " <td>0.428961</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.176875</td>\n", | |
| " <td>-0.712177</td>\n", | |
| " <td>-0.260505</td>\n", | |
| " <td>-0.008962</td>\n", | |
| " <td>0.120798</td>\n", | |
| " <td>0.156697</td>\n", | |
| " <td>0.123817</td>\n", | |
| " <td>-0.069584</td>\n", | |
| " <td>-0.631081</td>\n", | |
| " <td>-0.503107</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>pedigree</th>\n", | |
| " <td>-0.183650</td>\n", | |
| " <td>-0.415794</td>\n", | |
| " <td>-0.849095</td>\n", | |
| " <td>-0.422934</td>\n", | |
| " <td>-0.163478</td>\n", | |
| " <td>2.494999e-01</td>\n", | |
| " <td>0.550734</td>\n", | |
| " <td>-0.201845</td>\n", | |
| " <td>0.445531</td>\n", | |
| " <td>0.090581</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.734479</td>\n", | |
| " <td>0.820641</td>\n", | |
| " <td>-0.138604</td>\n", | |
| " <td>0.645414</td>\n", | |
| " <td>0.334605</td>\n", | |
| " <td>0.331809</td>\n", | |
| " <td>-0.893086</td>\n", | |
| " <td>0.014228</td>\n", | |
| " <td>-0.050464</td>\n", | |
| " <td>-0.126028</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>presence</th>\n", | |
| " <td>0.042307</td>\n", | |
| " <td>0.373298</td>\n", | |
| " <td>0.318494</td>\n", | |
| " <td>-0.429378</td>\n", | |
| " <td>-0.133881</td>\n", | |
| " <td>-4.116624e-01</td>\n", | |
| " <td>0.413157</td>\n", | |
| " <td>-0.014635</td>\n", | |
| " <td>0.295439</td>\n", | |
| " <td>-0.146233</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.003480</td>\n", | |
| " <td>0.755789</td>\n", | |
| " <td>0.245210</td>\n", | |
| " <td>0.493319</td>\n", | |
| " <td>0.140914</td>\n", | |
| " <td>-0.535924</td>\n", | |
| " <td>0.232693</td>\n", | |
| " <td>-0.142808</td>\n", | |
| " <td>0.555902</td>\n", | |
| " <td>-0.291969</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>due</th>\n", | |
| " <td>-0.082739</td>\n", | |
| " <td>-0.236547</td>\n", | |
| " <td>-0.583549</td>\n", | |
| " <td>-0.207350</td>\n", | |
| " <td>-0.562443</td>\n", | |
| " <td>3.145103e-01</td>\n", | |
| " <td>0.424465</td>\n", | |
| " <td>0.563453</td>\n", | |
| " <td>-0.335139</td>\n", | |
| " <td>0.763761</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.286914</td>\n", | |
| " <td>0.328274</td>\n", | |
| " <td>-0.167101</td>\n", | |
| " <td>0.537883</td>\n", | |
| " <td>0.462834</td>\n", | |
| " <td>-0.246100</td>\n", | |
| " <td>0.389896</td>\n", | |
| " <td>-0.395062</td>\n", | |
| " <td>0.338876</td>\n", | |
| " <td>-0.196595</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>mg_ndc</th>\n", | |
| " <td>0.254583</td>\n", | |
| " <td>0.118902</td>\n", | |
| " <td>-0.397409</td>\n", | |
| " <td>-0.555786</td>\n", | |
| " <td>0.064182</td>\n", | |
| " <td>-1.406718e-02</td>\n", | |
| " <td>0.255781</td>\n", | |
| " <td>-0.339742</td>\n", | |
| " <td>0.549717</td>\n", | |
| " <td>0.109165</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.547276</td>\n", | |
| " <td>0.697355</td>\n", | |
| " <td>0.636886</td>\n", | |
| " <td>-0.208276</td>\n", | |
| " <td>0.254340</td>\n", | |
| " <td>0.625140</td>\n", | |
| " <td>-0.849242</td>\n", | |
| " <td>0.208186</td>\n", | |
| " <td>0.082466</td>\n", | |
| " <td>0.105004</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>mg</th>\n", | |
| " <td>0.180624</td>\n", | |
| " <td>0.238867</td>\n", | |
| " <td>-0.245773</td>\n", | |
| " <td>-0.370714</td>\n", | |
| " <td>-0.008355</td>\n", | |
| " <td>-1.265083e-02</td>\n", | |
| " <td>0.360007</td>\n", | |
| " <td>0.158293</td>\n", | |
| " <td>0.216545</td>\n", | |
| " <td>0.104351</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.503235</td>\n", | |
| " <td>0.474098</td>\n", | |
| " <td>0.013973</td>\n", | |
| " <td>0.507388</td>\n", | |
| " <td>-0.040523</td>\n", | |
| " <td>0.731755</td>\n", | |
| " <td>-0.635973</td>\n", | |
| " <td>0.035107</td>\n", | |
| " <td>-0.005107</td>\n", | |
| " <td>0.035497</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>cross_contamination</th>\n", | |
| " <td>0.546137</td>\n", | |
| " <td>-0.165126</td>\n", | |
| " <td>0.748970</td>\n", | |
| " <td>-0.576038</td>\n", | |
| " <td>-0.646507</td>\n", | |
| " <td>-5.895789e-01</td>\n", | |
| " <td>0.050646</td>\n", | |
| " <td>0.926762</td>\n", | |
| " <td>0.030429</td>\n", | |
| " <td>0.367874</td>\n", | |
| " <td>...</td>\n", | |
| " <td>1.404816</td>\n", | |
| " <td>1.110056</td>\n", | |
| " <td>-0.790472</td>\n", | |
| " <td>0.762967</td>\n", | |
| " <td>0.492521</td>\n", | |
| " <td>0.127917</td>\n", | |
| " <td>-0.376043</td>\n", | |
| " <td>0.157067</td>\n", | |
| " <td>0.389089</td>\n", | |
| " <td>0.247304</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>recall_because_they</th>\n", | |
| " <td>0.090291</td>\n", | |
| " <td>-0.377112</td>\n", | |
| " <td>0.097433</td>\n", | |
| " <td>0.054534</td>\n", | |
| " <td>-0.034999</td>\n", | |
| " <td>-4.260801e-08</td>\n", | |
| " <td>0.031403</td>\n", | |
| " <td>0.647524</td>\n", | |
| " <td>-0.031235</td>\n", | |
| " <td>0.014662</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.490120</td>\n", | |
| " <td>-0.015589</td>\n", | |
| " <td>-0.156260</td>\n", | |
| " <td>-0.652013</td>\n", | |
| " <td>-0.023428</td>\n", | |
| " <td>0.377424</td>\n", | |
| " <td>0.086698</td>\n", | |
| " <td>0.022968</td>\n", | |
| " <td>-0.190156</td>\n", | |
| " <td>-0.776980</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>may</th>\n", | |
| " <td>0.429841</td>\n", | |
| " <td>-0.000277</td>\n", | |
| " <td>-0.076886</td>\n", | |
| " <td>-0.369745</td>\n", | |
| " <td>-0.164456</td>\n", | |
| " <td>-3.018114e-01</td>\n", | |
| " <td>0.611168</td>\n", | |
| " <td>0.061855</td>\n", | |
| " <td>-0.045191</td>\n", | |
| " <td>-0.123433</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.543194</td>\n", | |
| " <td>0.572797</td>\n", | |
| " <td>-0.129499</td>\n", | |
| " <td>0.608830</td>\n", | |
| " <td>0.224593</td>\n", | |
| " <td>0.096305</td>\n", | |
| " <td>0.180668</td>\n", | |
| " <td>0.100313</td>\n", | |
| " <td>0.003281</td>\n", | |
| " <td>0.028236</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>quality</th>\n", | |
| " <td>0.587960</td>\n", | |
| " <td>-0.820686</td>\n", | |
| " <td>-1.188513</td>\n", | |
| " <td>-0.743769</td>\n", | |
| " <td>-0.458716</td>\n", | |
| " <td>2.538852e-01</td>\n", | |
| " <td>0.028759</td>\n", | |
| " <td>-0.456984</td>\n", | |
| " <td>-0.288988</td>\n", | |
| " <td>0.847385</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.567179</td>\n", | |
| " <td>0.439567</td>\n", | |
| " <td>-0.077482</td>\n", | |
| " <td>-0.246606</td>\n", | |
| " <td>0.591101</td>\n", | |
| " <td>-0.848754</td>\n", | |
| " <td>0.629272</td>\n", | |
| " <td>-0.718522</td>\n", | |
| " <td>1.154461</td>\n", | |
| " <td>-1.393663</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>facility</th>\n", | |
| " <td>-0.051065</td>\n", | |
| " <td>0.055970</td>\n", | |
| " <td>-0.084826</td>\n", | |
| " <td>-0.205367</td>\n", | |
| " <td>0.693129</td>\n", | |
| " <td>-2.390375e-01</td>\n", | |
| " <td>0.239817</td>\n", | |
| " <td>0.345122</td>\n", | |
| " <td>0.093184</td>\n", | |
| " <td>0.691959</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.161289</td>\n", | |
| " <td>0.584826</td>\n", | |
| " <td>-0.922316</td>\n", | |
| " <td>-0.313895</td>\n", | |
| " <td>0.485026</td>\n", | |
| " <td>0.067188</td>\n", | |
| " <td>-0.314685</td>\n", | |
| " <td>-0.125226</td>\n", | |
| " <td>-0.161432</td>\n", | |
| " <td>-0.733695</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>02/12/15</th>\n", | |
| " <td>0.635095</td>\n", | |
| " <td>0.073688</td>\n", | |
| " <td>-0.018945</td>\n", | |
| " <td>-0.251970</td>\n", | |
| " <td>0.109926</td>\n", | |
| " <td>1.705191e-02</td>\n", | |
| " <td>-0.029990</td>\n", | |
| " <td>-0.180634</td>\n", | |
| " <td>-0.003162</td>\n", | |
| " <td>0.043237</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.534545</td>\n", | |
| " <td>-0.099753</td>\n", | |
| " <td>-0.588152</td>\n", | |
| " <td>-0.044220</td>\n", | |
| " <td>0.424661</td>\n", | |
| " <td>-0.528969</td>\n", | |
| " <td>-0.073393</td>\n", | |
| " <td>0.023062</td>\n", | |
| " <td>0.240035</td>\n", | |
| " <td>-0.667557</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>distribute_between_01/05/12</th>\n", | |
| " <td>0.487791</td>\n", | |
| " <td>0.119183</td>\n", | |
| " <td>0.088164</td>\n", | |
| " <td>-0.428254</td>\n", | |
| " <td>0.038100</td>\n", | |
| " <td>-1.564848e-01</td>\n", | |
| " <td>-0.050939</td>\n", | |
| " <td>0.282968</td>\n", | |
| " <td>0.101198</td>\n", | |
| " <td>0.515868</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.431443</td>\n", | |
| " <td>-0.061192</td>\n", | |
| " <td>-0.462266</td>\n", | |
| " <td>-0.301673</td>\n", | |
| " <td>0.158960</td>\n", | |
| " <td>0.202878</td>\n", | |
| " <td>0.032444</td>\n", | |
| " <td>0.106908</td>\n", | |
| " <td>0.141060</td>\n", | |
| " <td>-0.854404</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>...</th>\n", | |
| " <td>...</td>\n", | |
| " <td>...</td>\n", | |
| " <td>...</td>\n", | |
| " <td>...</td>\n", | |
| " <td>...</td>\n", | |
| " <td>...</td>\n", | |
| " <td>...</td>\n", | |
| " <td>...</td>\n", | |
| " <td>...</td>\n", | |
| " <td>...</td>\n", | |
| " <td>...</td>\n", | |
| " <td>...</td>\n", | |
| " <td>...</td>\n", | |
| " <td>...</td>\n", | |
| " <td>...</td>\n", | |
| " <td>...</td>\n", | |
| " <td>...</td>\n", | |
| " <td>...</td>\n", | |
| " <td>...</td>\n", | |
| " <td>...</td>\n", | |
| " <td>...</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>exp_6/6/2014</th>\n", | |
| " <td>0.123732</td>\n", | |
| " <td>0.260373</td>\n", | |
| " <td>-0.502664</td>\n", | |
| " <td>-0.201963</td>\n", | |
| " <td>-0.178212</td>\n", | |
| " <td>-3.814551e-01</td>\n", | |
| " <td>0.451382</td>\n", | |
| " <td>-0.462614</td>\n", | |
| " <td>-0.016023</td>\n", | |
| " <td>0.085736</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.671953</td>\n", | |
| " <td>0.805070</td>\n", | |
| " <td>-0.068457</td>\n", | |
| " <td>0.540329</td>\n", | |
| " <td>0.581324</td>\n", | |
| " <td>0.702510</td>\n", | |
| " <td>-0.406129</td>\n", | |
| " <td>0.038623</td>\n", | |
| " <td>-0.038386</td>\n", | |
| " <td>-0.328455</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>exp_6/25/2014</th>\n", | |
| " <td>0.244186</td>\n", | |
| " <td>-0.098127</td>\n", | |
| " <td>-0.281454</td>\n", | |
| " <td>-0.342072</td>\n", | |
| " <td>0.060017</td>\n", | |
| " <td>-2.112823e-01</td>\n", | |
| " <td>0.192961</td>\n", | |
| " <td>-0.559163</td>\n", | |
| " <td>-0.206508</td>\n", | |
| " <td>-0.279455</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.611520</td>\n", | |
| " <td>0.734607</td>\n", | |
| " <td>0.059976</td>\n", | |
| " <td>0.348866</td>\n", | |
| " <td>0.646266</td>\n", | |
| " <td>0.717060</td>\n", | |
| " <td>-0.492291</td>\n", | |
| " <td>-0.359080</td>\n", | |
| " <td>0.233809</td>\n", | |
| " <td>-0.572923</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>stability_datum_do_not</th>\n", | |
| " <td>0.049356</td>\n", | |
| " <td>-0.255431</td>\n", | |
| " <td>-0.407510</td>\n", | |
| " <td>-0.169126</td>\n", | |
| " <td>-0.054683</td>\n", | |
| " <td>-3.287490e-01</td>\n", | |
| " <td>0.058500</td>\n", | |
| " <td>-1.020380</td>\n", | |
| " <td>0.368327</td>\n", | |
| " <td>0.774952</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.460111</td>\n", | |
| " <td>0.188657</td>\n", | |
| " <td>0.365806</td>\n", | |
| " <td>-0.349524</td>\n", | |
| " <td>0.194516</td>\n", | |
| " <td>0.359418</td>\n", | |
| " <td>-0.437779</td>\n", | |
| " <td>0.079897</td>\n", | |
| " <td>0.159291</td>\n", | |
| " <td>-0.514144</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>precipitate</th>\n", | |
| " <td>-0.068525</td>\n", | |
| " <td>0.033515</td>\n", | |
| " <td>-0.086092</td>\n", | |
| " <td>-0.413860</td>\n", | |
| " <td>-0.120369</td>\n", | |
| " <td>-1.444217e-01</td>\n", | |
| " <td>-0.312695</td>\n", | |
| " <td>-0.079684</td>\n", | |
| " <td>-0.487023</td>\n", | |
| " <td>0.441104</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.166431</td>\n", | |
| " <td>0.077672</td>\n", | |
| " <td>0.176044</td>\n", | |
| " <td>-0.085269</td>\n", | |
| " <td>-0.121363</td>\n", | |
| " <td>0.945627</td>\n", | |
| " <td>-0.328054</td>\n", | |
| " <td>-0.555913</td>\n", | |
| " <td>-0.043336</td>\n", | |
| " <td>-0.336172</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>propranolol_hcl</th>\n", | |
| " <td>-0.453735</td>\n", | |
| " <td>-0.284003</td>\n", | |
| " <td>-0.855451</td>\n", | |
| " <td>-0.547269</td>\n", | |
| " <td>0.080355</td>\n", | |
| " <td>2.679408e-02</td>\n", | |
| " <td>0.150247</td>\n", | |
| " <td>-0.267999</td>\n", | |
| " <td>-0.086719</td>\n", | |
| " <td>0.150892</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.753667</td>\n", | |
| " <td>0.499701</td>\n", | |
| " <td>-0.210794</td>\n", | |
| " <td>0.384330</td>\n", | |
| " <td>0.271781</td>\n", | |
| " <td>0.706757</td>\n", | |
| " <td>-0.971606</td>\n", | |
| " <td>-0.094029</td>\n", | |
| " <td>0.472163</td>\n", | |
| " <td>-0.350248</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>multivitamin/multimineral</th>\n", | |
| " <td>0.417221</td>\n", | |
| " <td>-0.025444</td>\n", | |
| " <td>-0.293377</td>\n", | |
| " <td>0.149909</td>\n", | |
| " <td>0.223683</td>\n", | |
| " <td>1.361338e-01</td>\n", | |
| " <td>0.414827</td>\n", | |
| " <td>-0.390778</td>\n", | |
| " <td>-0.401109</td>\n", | |
| " <td>-0.052547</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.628714</td>\n", | |
| " <td>0.620430</td>\n", | |
| " <td>-0.242618</td>\n", | |
| " <td>0.506193</td>\n", | |
| " <td>0.412268</td>\n", | |
| " <td>0.755777</td>\n", | |
| " <td>-0.950551</td>\n", | |
| " <td>0.286023</td>\n", | |
| " <td>0.243958</td>\n", | |
| " <td>-0.134040</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>break</th>\n", | |
| " <td>-0.318687</td>\n", | |
| " <td>0.187491</td>\n", | |
| " <td>-0.315601</td>\n", | |
| " <td>0.078676</td>\n", | |
| " <td>-0.087504</td>\n", | |
| " <td>4.679605e-01</td>\n", | |
| " <td>0.567400</td>\n", | |
| " <td>0.272329</td>\n", | |
| " <td>-0.691238</td>\n", | |
| " <td>0.118210</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.524726</td>\n", | |
| " <td>0.291206</td>\n", | |
| " <td>-0.034012</td>\n", | |
| " <td>0.350973</td>\n", | |
| " <td>-0.255148</td>\n", | |
| " <td>-0.001081</td>\n", | |
| " <td>-1.175425</td>\n", | |
| " <td>-0.038030</td>\n", | |
| " <td>0.217955</td>\n", | |
| " <td>-0.392921</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>stability_failure</th>\n", | |
| " <td>0.128610</td>\n", | |
| " <td>-0.188772</td>\n", | |
| " <td>0.133110</td>\n", | |
| " <td>-0.685989</td>\n", | |
| " <td>0.097719</td>\n", | |
| " <td>1.092844e+00</td>\n", | |
| " <td>-0.513215</td>\n", | |
| " <td>-0.055847</td>\n", | |
| " <td>0.342174</td>\n", | |
| " <td>0.611064</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.366238</td>\n", | |
| " <td>0.172792</td>\n", | |
| " <td>0.359987</td>\n", | |
| " <td>-0.277620</td>\n", | |
| " <td>-0.480863</td>\n", | |
| " <td>0.743065</td>\n", | |
| " <td>-0.382206</td>\n", | |
| " <td>-0.209747</td>\n", | |
| " <td>0.431726</td>\n", | |
| " <td>-0.799726</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>preservative</th>\n", | |
| " <td>0.055042</td>\n", | |
| " <td>0.588597</td>\n", | |
| " <td>-0.479355</td>\n", | |
| " <td>-0.453066</td>\n", | |
| " <td>0.310705</td>\n", | |
| " <td>3.839197e-01</td>\n", | |
| " <td>0.644708</td>\n", | |
| " <td>-0.360716</td>\n", | |
| " <td>-0.355660</td>\n", | |
| " <td>0.855263</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.197735</td>\n", | |
| " <td>0.584663</td>\n", | |
| " <td>0.121469</td>\n", | |
| " <td>0.303326</td>\n", | |
| " <td>0.085117</td>\n", | |
| " <td>-0.005443</td>\n", | |
| " <td>-0.402300</td>\n", | |
| " <td>0.329036</td>\n", | |
| " <td>-0.382790</td>\n", | |
| " <td>-0.156805</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>stainless_steel</th>\n", | |
| " <td>0.053390</td>\n", | |
| " <td>-0.174206</td>\n", | |
| " <td>0.265159</td>\n", | |
| " <td>-0.813737</td>\n", | |
| " <td>-0.259793</td>\n", | |
| " <td>5.639494e-02</td>\n", | |
| " <td>-0.126170</td>\n", | |
| " <td>0.552584</td>\n", | |
| " <td>-0.513198</td>\n", | |
| " <td>-0.152963</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.758225</td>\n", | |
| " <td>0.963554</td>\n", | |
| " <td>0.276743</td>\n", | |
| " <td>0.540535</td>\n", | |
| " <td>-0.150424</td>\n", | |
| " <td>0.124394</td>\n", | |
| " <td>-0.597058</td>\n", | |
| " <td>0.036565</td>\n", | |
| " <td>0.143074</td>\n", | |
| " <td>-0.331011</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>do_not</th>\n", | |
| " <td>0.187873</td>\n", | |
| " <td>-0.152088</td>\n", | |
| " <td>-0.472893</td>\n", | |
| " <td>-0.302787</td>\n", | |
| " <td>-0.150021</td>\n", | |
| " <td>2.374495e-01</td>\n", | |
| " <td>0.380434</td>\n", | |
| " <td>0.019086</td>\n", | |
| " <td>-0.304325</td>\n", | |
| " <td>0.985389</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.035308</td>\n", | |
| " <td>0.392217</td>\n", | |
| " <td>0.016973</td>\n", | |
| " <td>0.350163</td>\n", | |
| " <td>0.054419</td>\n", | |
| " <td>0.383941</td>\n", | |
| " <td>0.063834</td>\n", | |
| " <td>0.500655</td>\n", | |
| " <td>-0.475887</td>\n", | |
| " <td>-0.417157</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>good_manufacturing_practices</th>\n", | |
| " <td>-0.204730</td>\n", | |
| " <td>-0.272180</td>\n", | |
| " <td>0.386396</td>\n", | |
| " <td>-0.126478</td>\n", | |
| " <td>0.037189</td>\n", | |
| " <td>-7.163196e-01</td>\n", | |
| " <td>0.060065</td>\n", | |
| " <td>0.658614</td>\n", | |
| " <td>-0.776333</td>\n", | |
| " <td>0.221561</td>\n", | |
| " <td>...</td>\n", | |
| " <td>-0.319302</td>\n", | |
| " <td>0.272429</td>\n", | |
| " <td>-0.044322</td>\n", | |
| " <td>0.843428</td>\n", | |
| " <td>0.257539</td>\n", | |
| " <td>-0.003327</td>\n", | |
| " <td>0.267934</td>\n", | |
| " <td>0.462099</td>\n", | |
| " <td>-0.688051</td>\n", | |
| " <td>-0.695076</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>calcium</th>\n", | |
| " <td>0.109527</td>\n", | |
| " <td>-0.041752</td>\n", | |
| " <td>-0.327194</td>\n", | |
| " <td>-0.339362</td>\n", | |
| " <td>-0.252808</td>\n", | |
| " <td>3.326128e-01</td>\n", | |
| " <td>0.181127</td>\n", | |
| " <td>0.115128</td>\n", | |
| " <td>0.235976</td>\n", | |
| " <td>1.218738</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.603660</td>\n", | |
| " <td>0.694440</td>\n", | |
| " <td>0.153814</td>\n", | |
| " <td>0.544600</td>\n", | |
| " <td>0.299468</td>\n", | |
| " <td>0.412340</td>\n", | |
| " <td>-1.166201</td>\n", | |
| " <td>-0.020409</td>\n", | |
| " <td>0.199746</td>\n", | |
| " <td>-0.273912</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>discoloration</th>\n", | |
| " <td>0.002643</td>\n", | |
| " <td>0.462822</td>\n", | |
| " <td>-0.163611</td>\n", | |
| " <td>-0.311926</td>\n", | |
| " <td>-0.586656</td>\n", | |
| " <td>-4.881225e-01</td>\n", | |
| " <td>0.024960</td>\n", | |
| " <td>0.315812</td>\n", | |
| " <td>0.184850</td>\n", | |
| " <td>0.027123</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.142227</td>\n", | |
| " <td>0.570476</td>\n", | |
| " <td>-0.397453</td>\n", | |
| " <td>-0.115908</td>\n", | |
| " <td>0.058452</td>\n", | |
| " <td>0.682327</td>\n", | |
| " <td>-0.359661</td>\n", | |
| " <td>0.497713</td>\n", | |
| " <td>-0.077592</td>\n", | |
| " <td>-0.187794</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>approve_nda/anda</th>\n", | |
| " <td>-0.165780</td>\n", | |
| " <td>-0.203468</td>\n", | |
| " <td>-0.129073</td>\n", | |
| " <td>0.101168</td>\n", | |
| " <td>-0.233593</td>\n", | |
| " <td>-1.045111e-01</td>\n", | |
| " <td>0.231025</td>\n", | |
| " <td>-0.585117</td>\n", | |
| " <td>0.255553</td>\n", | |
| " <td>-0.249335</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.784463</td>\n", | |
| " <td>0.008002</td>\n", | |
| " <td>0.324189</td>\n", | |
| " <td>-0.094483</td>\n", | |
| " <td>0.583553</td>\n", | |
| " <td>0.110497</td>\n", | |
| " <td>0.193130</td>\n", | |
| " <td>-0.156360</td>\n", | |
| " <td>0.187089</td>\n", | |
| " <td>-0.964792</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>substance</th>\n", | |
| " <td>0.083305</td>\n", | |
| " <td>-0.193139</td>\n", | |
| " <td>0.090817</td>\n", | |
| " <td>-0.290849</td>\n", | |
| " <td>-0.359260</td>\n", | |
| " <td>-6.449277e-02</td>\n", | |
| " <td>0.593015</td>\n", | |
| " <td>-0.158613</td>\n", | |
| " <td>-0.043727</td>\n", | |
| " <td>0.047042</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.686860</td>\n", | |
| " <td>0.566097</td>\n", | |
| " <td>-0.366434</td>\n", | |
| " <td>-0.060759</td>\n", | |
| " <td>0.059307</td>\n", | |
| " <td>0.049056</td>\n", | |
| " <td>0.058596</td>\n", | |
| " <td>0.254278</td>\n", | |
| " <td>-0.088240</td>\n", | |
| " <td>-0.340175</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>insufficient_datum</th>\n", | |
| " <td>0.071994</td>\n", | |
| " <td>-0.192453</td>\n", | |
| " <td>0.077260</td>\n", | |
| " <td>-0.731172</td>\n", | |
| " <td>0.108884</td>\n", | |
| " <td>1.257619e+00</td>\n", | |
| " <td>-0.408081</td>\n", | |
| " <td>0.187564</td>\n", | |
| " <td>0.272687</td>\n", | |
| " <td>0.535653</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.501408</td>\n", | |
| " <td>0.068721</td>\n", | |
| " <td>0.452180</td>\n", | |
| " <td>-0.211653</td>\n", | |
| " <td>-0.494282</td>\n", | |
| " <td>0.595109</td>\n", | |
| " <td>-0.310056</td>\n", | |
| " <td>0.064565</td>\n", | |
| " <td>0.471530</td>\n", | |
| " <td>-0.737062</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>dr</th>\n", | |
| " <td>0.091955</td>\n", | |
| " <td>-0.888579</td>\n", | |
| " <td>-0.846769</td>\n", | |
| " <td>-0.501187</td>\n", | |
| " <td>-0.431423</td>\n", | |
| " <td>1.355121e-01</td>\n", | |
| " <td>0.257163</td>\n", | |
| " <td>-0.263879</td>\n", | |
| " <td>0.102050</td>\n", | |
| " <td>-0.226927</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.506841</td>\n", | |
| " <td>0.650217</td>\n", | |
| " <td>-0.068841</td>\n", | |
| " <td>0.551029</td>\n", | |
| " <td>0.457740</td>\n", | |
| " <td>0.564364</td>\n", | |
| " <td>-0.939611</td>\n", | |
| " <td>0.338181</td>\n", | |
| " <td>0.279912</td>\n", | |
| " <td>-0.783463</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>salicylic_acid</th>\n", | |
| " <td>-0.195229</td>\n", | |
| " <td>-0.520733</td>\n", | |
| " <td>0.209155</td>\n", | |
| " <td>-0.252709</td>\n", | |
| " <td>0.749793</td>\n", | |
| " <td>3.344107e-01</td>\n", | |
| " <td>0.521556</td>\n", | |
| " <td>-0.372945</td>\n", | |
| " <td>-0.025462</td>\n", | |
| " <td>-0.604147</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.624181</td>\n", | |
| " <td>0.050387</td>\n", | |
| " <td>-0.023391</td>\n", | |
| " <td>0.131366</td>\n", | |
| " <td>0.421586</td>\n", | |
| " <td>0.224686</td>\n", | |
| " <td>-0.071084</td>\n", | |
| " <td>0.177473</td>\n", | |
| " <td>0.115413</td>\n", | |
| " <td>-0.940842</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>stability_time_point</th>\n", | |
| " <td>-0.159436</td>\n", | |
| " <td>-0.235678</td>\n", | |
| " <td>-0.616431</td>\n", | |
| " <td>-0.478709</td>\n", | |
| " <td>0.671741</td>\n", | |
| " <td>3.512335e-01</td>\n", | |
| " <td>0.235527</td>\n", | |
| " <td>-0.398954</td>\n", | |
| " <td>-0.302937</td>\n", | |
| " <td>0.291169</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.153095</td>\n", | |
| " <td>-0.302260</td>\n", | |
| " <td>-0.045905</td>\n", | |
| " <td>0.337051</td>\n", | |
| " <td>0.115155</td>\n", | |
| " <td>0.355597</td>\n", | |
| " <td>-0.358764</td>\n", | |
| " <td>-0.141571</td>\n", | |
| " <td>0.444522</td>\n", | |
| " <td>-1.462083</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>hence</th>\n", | |
| " <td>0.210536</td>\n", | |
| " <td>0.110444</td>\n", | |
| " <td>-0.364460</td>\n", | |
| " <td>0.257135</td>\n", | |
| " <td>0.075636</td>\n", | |
| " <td>2.398437e-01</td>\n", | |
| " <td>0.038534</td>\n", | |
| " <td>0.264866</td>\n", | |
| " <td>0.674395</td>\n", | |
| " <td>-0.088549</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.266611</td>\n", | |
| " <td>0.809901</td>\n", | |
| " <td>-0.301491</td>\n", | |
| " <td>-0.028232</td>\n", | |
| " <td>0.252878</td>\n", | |
| " <td>0.051660</td>\n", | |
| " <td>-0.016800</td>\n", | |
| " <td>-0.362617</td>\n", | |
| " <td>0.699495</td>\n", | |
| " <td>-0.971199</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>subpotent_single_ingredient</th>\n", | |
| " <td>0.242071</td>\n", | |
| " <td>-0.251082</td>\n", | |
| " <td>-0.436063</td>\n", | |
| " <td>-0.373570</td>\n", | |
| " <td>0.089401</td>\n", | |
| " <td>-3.989760e-02</td>\n", | |
| " <td>0.153367</td>\n", | |
| " <td>-0.083442</td>\n", | |
| " <td>-0.552353</td>\n", | |
| " <td>0.309919</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.654329</td>\n", | |
| " <td>0.038740</td>\n", | |
| " <td>-0.716861</td>\n", | |
| " <td>0.435336</td>\n", | |
| " <td>0.349279</td>\n", | |
| " <td>0.618122</td>\n", | |
| " <td>-0.182675</td>\n", | |
| " <td>0.425007</td>\n", | |
| " <td>-0.030217</td>\n", | |
| " <td>-0.608149</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>lozenge</th>\n", | |
| " <td>-0.146890</td>\n", | |
| " <td>-0.910858</td>\n", | |
| " <td>-0.415331</td>\n", | |
| " <td>-0.162915</td>\n", | |
| " <td>0.292086</td>\n", | |
| " <td>-1.196768e-01</td>\n", | |
| " <td>0.156596</td>\n", | |
| " <td>-0.441530</td>\n", | |
| " <td>0.247628</td>\n", | |
| " <td>0.107503</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.348825</td>\n", | |
| " <td>0.297437</td>\n", | |
| " <td>-0.325696</td>\n", | |
| " <td>0.115757</td>\n", | |
| " <td>0.026851</td>\n", | |
| " <td>0.231624</td>\n", | |
| " <td>-0.345866</td>\n", | |
| " <td>0.419189</td>\n", | |
| " <td>-0.216863</td>\n", | |
| " <td>-0.675952</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>previous_lot_there</th>\n", | |
| " <td>0.103645</td>\n", | |
| " <td>-0.170941</td>\n", | |
| " <td>0.112203</td>\n", | |
| " <td>-0.732700</td>\n", | |
| " <td>0.085244</td>\n", | |
| " <td>1.212467e+00</td>\n", | |
| " <td>-0.474048</td>\n", | |
| " <td>0.051048</td>\n", | |
| " <td>0.392926</td>\n", | |
| " <td>0.527287</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.439697</td>\n", | |
| " <td>0.107292</td>\n", | |
| " <td>0.420130</td>\n", | |
| " <td>-0.265406</td>\n", | |
| " <td>-0.533381</td>\n", | |
| " <td>0.699781</td>\n", | |
| " <td>-0.331206</td>\n", | |
| " <td>-0.059230</td>\n", | |
| " <td>0.433114</td>\n", | |
| " <td>-0.782535</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>determine_that_other</th>\n", | |
| " <td>0.033456</td>\n", | |
| " <td>-0.267150</td>\n", | |
| " <td>0.025440</td>\n", | |
| " <td>-0.800761</td>\n", | |
| " <td>0.103453</td>\n", | |
| " <td>1.213115e+00</td>\n", | |
| " <td>-0.389119</td>\n", | |
| " <td>0.222537</td>\n", | |
| " <td>0.177603</td>\n", | |
| " <td>0.435548</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.466691</td>\n", | |
| " <td>-0.008785</td>\n", | |
| " <td>0.432847</td>\n", | |
| " <td>-0.190846</td>\n", | |
| " <td>-0.468886</td>\n", | |
| " <td>0.472048</td>\n", | |
| " <td>-0.324222</td>\n", | |
| " <td>0.156423</td>\n", | |
| " <td>0.405998</td>\n", | |
| " <td>-0.654671</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>burkholderia_cepacia</th>\n", | |
| " <td>0.425895</td>\n", | |
| " <td>-0.225407</td>\n", | |
| " <td>0.219679</td>\n", | |
| " <td>-0.909065</td>\n", | |
| " <td>-0.420190</td>\n", | |
| " <td>5.274637e-01</td>\n", | |
| " <td>0.308330</td>\n", | |
| " <td>1.012187</td>\n", | |
| " <td>-0.541867</td>\n", | |
| " <td>0.014436</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.955094</td>\n", | |
| " <td>1.087692</td>\n", | |
| " <td>-0.327379</td>\n", | |
| " <td>0.203308</td>\n", | |
| " <td>0.325728</td>\n", | |
| " <td>-0.273550</td>\n", | |
| " <td>0.297474</td>\n", | |
| " <td>0.830083</td>\n", | |
| " <td>-0.312336</td>\n", | |
| " <td>0.019949</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>market_without</th>\n", | |
| " <td>-0.058764</td>\n", | |
| " <td>-0.251950</td>\n", | |
| " <td>-0.085931</td>\n", | |
| " <td>0.141777</td>\n", | |
| " <td>-0.221918</td>\n", | |
| " <td>-1.816617e-01</td>\n", | |
| " <td>0.495476</td>\n", | |
| " <td>-0.568201</td>\n", | |
| " <td>0.342282</td>\n", | |
| " <td>-0.253555</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.694742</td>\n", | |
| " <td>0.055534</td>\n", | |
| " <td>0.111120</td>\n", | |
| " <td>-0.203184</td>\n", | |
| " <td>0.478466</td>\n", | |
| " <td>-0.062840</td>\n", | |
| " <td>0.127311</td>\n", | |
| " <td>-0.001967</td>\n", | |
| " <td>0.144946</td>\n", | |
| " <td>-0.978934</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>quality_review</th>\n", | |
| " <td>0.166578</td>\n", | |
| " <td>-0.083396</td>\n", | |
| " <td>0.174855</td>\n", | |
| " <td>-0.646447</td>\n", | |
| " <td>0.092615</td>\n", | |
| " <td>1.069835e+00</td>\n", | |
| " <td>-0.550002</td>\n", | |
| " <td>-0.192062</td>\n", | |
| " <td>0.437350</td>\n", | |
| " <td>0.618896</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.337255</td>\n", | |
| " <td>0.124640</td>\n", | |
| " <td>0.316804</td>\n", | |
| " <td>-0.235937</td>\n", | |
| " <td>-0.510923</td>\n", | |
| " <td>0.877442</td>\n", | |
| " <td>-0.289462</td>\n", | |
| " <td>-0.270052</td>\n", | |
| " <td>0.417941</td>\n", | |
| " <td>-0.803751</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>tadalafil</th>\n", | |
| " <td>-0.369522</td>\n", | |
| " <td>-0.067828</td>\n", | |
| " <td>0.261457</td>\n", | |
| " <td>0.127375</td>\n", | |
| " <td>-0.115995</td>\n", | |
| " <td>1.847755e-01</td>\n", | |
| " <td>-0.069203</td>\n", | |
| " <td>0.249611</td>\n", | |
| " <td>-0.423353</td>\n", | |
| " <td>-0.396357</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.439668</td>\n", | |
| " <td>0.320275</td>\n", | |
| " <td>-0.436805</td>\n", | |
| " <td>0.639454</td>\n", | |
| " <td>0.280085</td>\n", | |
| " <td>-0.091469</td>\n", | |
| " <td>-0.390900</td>\n", | |
| " <td>0.375969</td>\n", | |
| " <td>-0.062292</td>\n", | |
| " <td>-0.337352</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>manufacturing_firm</th>\n", | |
| " <td>0.121493</td>\n", | |
| " <td>-0.581098</td>\n", | |
| " <td>0.275406</td>\n", | |
| " <td>0.107986</td>\n", | |
| " <td>-0.208517</td>\n", | |
| " <td>-1.369090e-01</td>\n", | |
| " <td>0.347648</td>\n", | |
| " <td>0.587570</td>\n", | |
| " <td>0.203935</td>\n", | |
| " <td>0.759921</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.758731</td>\n", | |
| " <td>1.041262</td>\n", | |
| " <td>-0.871369</td>\n", | |
| " <td>0.395397</td>\n", | |
| " <td>-0.129529</td>\n", | |
| " <td>0.370408</td>\n", | |
| " <td>-0.257274</td>\n", | |
| " <td>0.303240</td>\n", | |
| " <td>-0.300379</td>\n", | |
| " <td>-0.106373</td>\n", | |
| " </tr>\n", | |
| " </tbody>\n", | |
| "</table>\n", | |
| "<p>422 rows × 100 columns</p>\n", | |
| "</div>" | |
| ], | |
| "text/plain": [ | |
| " 0 1 2 3 \\\n", | |
| "of 0.250408 -0.426319 -0.465205 -0.204381 \n", | |
| "be 0.349246 -0.090556 -0.084044 -0.466857 \n", | |
| "the 0.067424 0.080021 0.033621 0.182714 \n", | |
| "sterility 0.303488 -0.228153 -0.219394 -0.302188 \n", | |
| "assurance 0.240375 -0.008034 -0.594212 0.349680 \n", | |
| "product 0.167806 -0.021859 0.185118 -0.030679 \n", | |
| "lack 0.666243 -0.235590 -0.383695 0.450461 \n", | |
| "and 0.013336 0.060998 -0.056626 0.209782 \n", | |
| "in 0.106915 0.433992 -0.022223 -0.229789 \n", | |
| "to -0.055777 -0.142672 -0.052781 -0.160961 \n", | |
| "with 0.139916 0.056741 0.019701 -0.038652 \n", | |
| "a 0.515118 0.419556 0.291573 -0.048714 \n", | |
| "penicillin 0.331125 0.195782 -0.216822 -0.540945 \n", | |
| "all 0.767341 -0.377597 -0.033244 -0.079458 \n", | |
| "recall -0.068916 -0.640970 0.074110 -0.269826 \n", | |
| "tablet 0.150178 -0.334870 -0.514988 -0.383748 \n", | |
| "repackaged 0.234697 0.004605 -0.236921 -0.172754 \n", | |
| "lot 0.410613 0.485917 -0.436936 -0.594825 \n", | |
| "pedigree -0.183650 -0.415794 -0.849095 -0.422934 \n", | |
| "presence 0.042307 0.373298 0.318494 -0.429378 \n", | |
| "due -0.082739 -0.236547 -0.583549 -0.207350 \n", | |
| "mg_ndc 0.254583 0.118902 -0.397409 -0.555786 \n", | |
| "mg 0.180624 0.238867 -0.245773 -0.370714 \n", | |
| "cross_contamination 0.546137 -0.165126 0.748970 -0.576038 \n", | |
| "recall_because_they 0.090291 -0.377112 0.097433 0.054534 \n", | |
| "may 0.429841 -0.000277 -0.076886 -0.369745 \n", | |
| "quality 0.587960 -0.820686 -1.188513 -0.743769 \n", | |
| "facility -0.051065 0.055970 -0.084826 -0.205367 \n", | |
| "02/12/15 0.635095 0.073688 -0.018945 -0.251970 \n", | |
| "distribute_between_01/05/12 0.487791 0.119183 0.088164 -0.428254 \n", | |
| "... ... ... ... ... \n", | |
| "exp_6/6/2014 0.123732 0.260373 -0.502664 -0.201963 \n", | |
| "exp_6/25/2014 0.244186 -0.098127 -0.281454 -0.342072 \n", | |
| "stability_datum_do_not 0.049356 -0.255431 -0.407510 -0.169126 \n", | |
| "precipitate -0.068525 0.033515 -0.086092 -0.413860 \n", | |
| "propranolol_hcl -0.453735 -0.284003 -0.855451 -0.547269 \n", | |
| "multivitamin/multimineral 0.417221 -0.025444 -0.293377 0.149909 \n", | |
| "break -0.318687 0.187491 -0.315601 0.078676 \n", | |
| "stability_failure 0.128610 -0.188772 0.133110 -0.685989 \n", | |
| "preservative 0.055042 0.588597 -0.479355 -0.453066 \n", | |
| "stainless_steel 0.053390 -0.174206 0.265159 -0.813737 \n", | |
| "do_not 0.187873 -0.152088 -0.472893 -0.302787 \n", | |
| "good_manufacturing_practices -0.204730 -0.272180 0.386396 -0.126478 \n", | |
| "calcium 0.109527 -0.041752 -0.327194 -0.339362 \n", | |
| "discoloration 0.002643 0.462822 -0.163611 -0.311926 \n", | |
| "approve_nda/anda -0.165780 -0.203468 -0.129073 0.101168 \n", | |
| "substance 0.083305 -0.193139 0.090817 -0.290849 \n", | |
| "insufficient_datum 0.071994 -0.192453 0.077260 -0.731172 \n", | |
| "dr 0.091955 -0.888579 -0.846769 -0.501187 \n", | |
| "salicylic_acid -0.195229 -0.520733 0.209155 -0.252709 \n", | |
| "stability_time_point -0.159436 -0.235678 -0.616431 -0.478709 \n", | |
| "hence 0.210536 0.110444 -0.364460 0.257135 \n", | |
| "subpotent_single_ingredient 0.242071 -0.251082 -0.436063 -0.373570 \n", | |
| "lozenge -0.146890 -0.910858 -0.415331 -0.162915 \n", | |
| "previous_lot_there 0.103645 -0.170941 0.112203 -0.732700 \n", | |
| "determine_that_other 0.033456 -0.267150 0.025440 -0.800761 \n", | |
| "burkholderia_cepacia 0.425895 -0.225407 0.219679 -0.909065 \n", | |
| "market_without -0.058764 -0.251950 -0.085931 0.141777 \n", | |
| "quality_review 0.166578 -0.083396 0.174855 -0.646447 \n", | |
| "tadalafil -0.369522 -0.067828 0.261457 0.127375 \n", | |
| "manufacturing_firm 0.121493 -0.581098 0.275406 0.107986 \n", | |
| "\n", | |
| " 4 5 6 7 \\\n", | |
| "of -0.392508 2.501663e-01 -0.158044 0.304148 \n", | |
| "be -0.324749 -5.094894e-02 -0.038947 0.237301 \n", | |
| "the -0.021266 -5.362771e-01 0.272588 0.268369 \n", | |
| "sterility -0.070613 4.507700e-01 -0.125470 -0.174656 \n", | |
| "assurance -0.603484 5.307044e-01 0.029388 0.626848 \n", | |
| "product -0.188094 1.409636e-01 -0.239155 0.544782 \n", | |
| "lack -0.533766 6.104476e-01 -0.394387 0.135181 \n", | |
| "and 0.027815 2.330013e-02 -0.311109 0.436705 \n", | |
| "in -0.483802 -2.567068e-01 -0.353319 -0.128223 \n", | |
| "to -0.114285 9.478991e-02 0.197403 0.107659 \n", | |
| "with -0.131373 3.501720e-02 -0.514761 0.129861 \n", | |
| "a -0.854946 2.778567e-01 0.081357 0.085357 \n", | |
| "penicillin -0.070391 -1.685262e-01 0.119359 0.104435 \n", | |
| "all -0.263991 3.638779e-01 -0.942502 0.240766 \n", | |
| "recall -0.126685 1.882937e-01 0.419718 0.327580 \n", | |
| "tablet -0.048423 1.644191e-02 0.362565 -0.134630 \n", | |
| "repackaged -0.094025 -1.371086e-01 -0.204735 0.162600 \n", | |
| "lot 0.186103 2.725144e-01 -0.276863 -0.037303 \n", | |
| "pedigree -0.163478 2.494999e-01 0.550734 -0.201845 \n", | |
| "presence -0.133881 -4.116624e-01 0.413157 -0.014635 \n", | |
| "due -0.562443 3.145103e-01 0.424465 0.563453 \n", | |
| "mg_ndc 0.064182 -1.406718e-02 0.255781 -0.339742 \n", | |
| "mg -0.008355 -1.265083e-02 0.360007 0.158293 \n", | |
| "cross_contamination -0.646507 -5.895789e-01 0.050646 0.926762 \n", | |
| "recall_because_they -0.034999 -4.260801e-08 0.031403 0.647524 \n", | |
| "may -0.164456 -3.018114e-01 0.611168 0.061855 \n", | |
| "quality -0.458716 2.538852e-01 0.028759 -0.456984 \n", | |
| "facility 0.693129 -2.390375e-01 0.239817 0.345122 \n", | |
| "02/12/15 0.109926 1.705191e-02 -0.029990 -0.180634 \n", | |
| "distribute_between_01/05/12 0.038100 -1.564848e-01 -0.050939 0.282968 \n", | |
| "... ... ... ... ... \n", | |
| "exp_6/6/2014 -0.178212 -3.814551e-01 0.451382 -0.462614 \n", | |
| "exp_6/25/2014 0.060017 -2.112823e-01 0.192961 -0.559163 \n", | |
| "stability_datum_do_not -0.054683 -3.287490e-01 0.058500 -1.020380 \n", | |
| "precipitate -0.120369 -1.444217e-01 -0.312695 -0.079684 \n", | |
| "propranolol_hcl 0.080355 2.679408e-02 0.150247 -0.267999 \n", | |
| "multivitamin/multimineral 0.223683 1.361338e-01 0.414827 -0.390778 \n", | |
| "break -0.087504 4.679605e-01 0.567400 0.272329 \n", | |
| "stability_failure 0.097719 1.092844e+00 -0.513215 -0.055847 \n", | |
| "preservative 0.310705 3.839197e-01 0.644708 -0.360716 \n", | |
| "stainless_steel -0.259793 5.639494e-02 -0.126170 0.552584 \n", | |
| "do_not -0.150021 2.374495e-01 0.380434 0.019086 \n", | |
| "good_manufacturing_practices 0.037189 -7.163196e-01 0.060065 0.658614 \n", | |
| "calcium -0.252808 3.326128e-01 0.181127 0.115128 \n", | |
| "discoloration -0.586656 -4.881225e-01 0.024960 0.315812 \n", | |
| "approve_nda/anda -0.233593 -1.045111e-01 0.231025 -0.585117 \n", | |
| "substance -0.359260 -6.449277e-02 0.593015 -0.158613 \n", | |
| "insufficient_datum 0.108884 1.257619e+00 -0.408081 0.187564 \n", | |
| "dr -0.431423 1.355121e-01 0.257163 -0.263879 \n", | |
| "salicylic_acid 0.749793 3.344107e-01 0.521556 -0.372945 \n", | |
| "stability_time_point 0.671741 3.512335e-01 0.235527 -0.398954 \n", | |
| "hence 0.075636 2.398437e-01 0.038534 0.264866 \n", | |
| "subpotent_single_ingredient 0.089401 -3.989760e-02 0.153367 -0.083442 \n", | |
| "lozenge 0.292086 -1.196768e-01 0.156596 -0.441530 \n", | |
| "previous_lot_there 0.085244 1.212467e+00 -0.474048 0.051048 \n", | |
| "determine_that_other 0.103453 1.213115e+00 -0.389119 0.222537 \n", | |
| "burkholderia_cepacia -0.420190 5.274637e-01 0.308330 1.012187 \n", | |
| "market_without -0.221918 -1.816617e-01 0.495476 -0.568201 \n", | |
| "quality_review 0.092615 1.069835e+00 -0.550002 -0.192062 \n", | |
| "tadalafil -0.115995 1.847755e-01 -0.069203 0.249611 \n", | |
| "manufacturing_firm -0.208517 -1.369090e-01 0.347648 0.587570 \n", | |
| "\n", | |
| " 8 9 ... 90 \\\n", | |
| "of 0.218495 0.383973 ... -0.232480 \n", | |
| "be 0.152482 0.056053 ... -0.071386 \n", | |
| "the -0.154391 0.496174 ... 0.333724 \n", | |
| "sterility 0.334442 0.810358 ... 0.372313 \n", | |
| "assurance 0.348065 0.077353 ... 0.388538 \n", | |
| "product 0.352739 0.016653 ... 0.025098 \n", | |
| "lack -0.221998 0.284320 ... 0.096456 \n", | |
| "and -0.339965 0.205782 ... 0.457187 \n", | |
| "in -0.179633 0.151007 ... 0.287440 \n", | |
| "to 0.009553 0.110412 ... 0.030555 \n", | |
| "with 0.129339 0.307071 ... 0.361517 \n", | |
| "a 0.118027 0.006842 ... 0.433700 \n", | |
| "penicillin -0.079008 0.072145 ... 0.097845 \n", | |
| "all -0.142430 0.114177 ... 0.210978 \n", | |
| "recall 0.170750 0.496003 ... 0.107886 \n", | |
| "tablet -0.053598 -0.112347 ... 0.610540 \n", | |
| "repackaged -0.507803 0.215561 ... 0.202148 \n", | |
| "lot 0.059433 0.428961 ... 0.176875 \n", | |
| "pedigree 0.445531 0.090581 ... 0.734479 \n", | |
| "presence 0.295439 -0.146233 ... 0.003480 \n", | |
| "due -0.335139 0.763761 ... 0.286914 \n", | |
| "mg_ndc 0.549717 0.109165 ... 0.547276 \n", | |
| "mg 0.216545 0.104351 ... 0.503235 \n", | |
| "cross_contamination 0.030429 0.367874 ... 1.404816 \n", | |
| "recall_because_they -0.031235 0.014662 ... 0.490120 \n", | |
| "may -0.045191 -0.123433 ... 0.543194 \n", | |
| "quality -0.288988 0.847385 ... 0.567179 \n", | |
| "facility 0.093184 0.691959 ... 0.161289 \n", | |
| "02/12/15 -0.003162 0.043237 ... 0.534545 \n", | |
| "distribute_between_01/05/12 0.101198 0.515868 ... 0.431443 \n", | |
| "... ... ... ... ... \n", | |
| "exp_6/6/2014 -0.016023 0.085736 ... 0.671953 \n", | |
| "exp_6/25/2014 -0.206508 -0.279455 ... 0.611520 \n", | |
| "stability_datum_do_not 0.368327 0.774952 ... 0.460111 \n", | |
| "precipitate -0.487023 0.441104 ... 0.166431 \n", | |
| "propranolol_hcl -0.086719 0.150892 ... 0.753667 \n", | |
| "multivitamin/multimineral -0.401109 -0.052547 ... 0.628714 \n", | |
| "break -0.691238 0.118210 ... 0.524726 \n", | |
| "stability_failure 0.342174 0.611064 ... 0.366238 \n", | |
| "preservative -0.355660 0.855263 ... 0.197735 \n", | |
| "stainless_steel -0.513198 -0.152963 ... 0.758225 \n", | |
| "do_not -0.304325 0.985389 ... 0.035308 \n", | |
| "good_manufacturing_practices -0.776333 0.221561 ... -0.319302 \n", | |
| "calcium 0.235976 1.218738 ... 0.603660 \n", | |
| "discoloration 0.184850 0.027123 ... 0.142227 \n", | |
| "approve_nda/anda 0.255553 -0.249335 ... 0.784463 \n", | |
| "substance -0.043727 0.047042 ... 0.686860 \n", | |
| "insufficient_datum 0.272687 0.535653 ... 0.501408 \n", | |
| "dr 0.102050 -0.226927 ... 0.506841 \n", | |
| "salicylic_acid -0.025462 -0.604147 ... 0.624181 \n", | |
| "stability_time_point -0.302937 0.291169 ... 0.153095 \n", | |
| "hence 0.674395 -0.088549 ... 0.266611 \n", | |
| "subpotent_single_ingredient -0.552353 0.309919 ... 0.654329 \n", | |
| "lozenge 0.247628 0.107503 ... 0.348825 \n", | |
| "previous_lot_there 0.392926 0.527287 ... 0.439697 \n", | |
| "determine_that_other 0.177603 0.435548 ... 0.466691 \n", | |
| "burkholderia_cepacia -0.541867 0.014436 ... 0.955094 \n", | |
| "market_without 0.342282 -0.253555 ... 0.694742 \n", | |
| "quality_review 0.437350 0.618896 ... 0.337255 \n", | |
| "tadalafil -0.423353 -0.396357 ... 0.439668 \n", | |
| "manufacturing_firm 0.203935 0.759921 ... 0.758731 \n", | |
| "\n", | |
| " 91 92 93 94 \\\n", | |
| "of -0.333301 0.318591 0.029734 -0.192704 \n", | |
| "be 0.110380 -0.113990 0.206021 0.123648 \n", | |
| "the -0.015216 0.251310 0.469194 0.128453 \n", | |
| "sterility 0.377984 -0.309888 -0.746572 -0.342844 \n", | |
| "assurance 0.002734 -0.239271 -0.082077 -0.047349 \n", | |
| "product -0.186757 -0.083697 -0.093634 -0.106546 \n", | |
| "lack 0.144373 0.413397 -0.051289 -0.165963 \n", | |
| "and -0.042881 0.268263 -0.257201 0.311483 \n", | |
| "in -0.491665 0.040266 0.345051 0.493042 \n", | |
| "to 0.313960 0.226491 0.116636 0.144624 \n", | |
| "with 0.152685 -0.730674 0.113176 0.702688 \n", | |
| "a 0.406811 -0.138931 -0.100497 0.305594 \n", | |
| "penicillin 0.443677 -0.943778 -0.739152 0.490247 \n", | |
| "all 0.069528 -0.270203 -0.461119 0.401482 \n", | |
| "recall 0.658807 0.094854 0.475893 0.054919 \n", | |
| "tablet 0.415149 0.225299 0.186411 0.109859 \n", | |
| "repackaged 0.176992 -0.694359 -0.579576 0.460001 \n", | |
| "lot -0.712177 -0.260505 -0.008962 0.120798 \n", | |
| "pedigree 0.820641 -0.138604 0.645414 0.334605 \n", | |
| "presence 0.755789 0.245210 0.493319 0.140914 \n", | |
| "due 0.328274 -0.167101 0.537883 0.462834 \n", | |
| "mg_ndc 0.697355 0.636886 -0.208276 0.254340 \n", | |
| "mg 0.474098 0.013973 0.507388 -0.040523 \n", | |
| "cross_contamination 1.110056 -0.790472 0.762967 0.492521 \n", | |
| "recall_because_they -0.015589 -0.156260 -0.652013 -0.023428 \n", | |
| "may 0.572797 -0.129499 0.608830 0.224593 \n", | |
| "quality 0.439567 -0.077482 -0.246606 0.591101 \n", | |
| "facility 0.584826 -0.922316 -0.313895 0.485026 \n", | |
| "02/12/15 -0.099753 -0.588152 -0.044220 0.424661 \n", | |
| "distribute_between_01/05/12 -0.061192 -0.462266 -0.301673 0.158960 \n", | |
| "... ... ... ... ... \n", | |
| "exp_6/6/2014 0.805070 -0.068457 0.540329 0.581324 \n", | |
| "exp_6/25/2014 0.734607 0.059976 0.348866 0.646266 \n", | |
| "stability_datum_do_not 0.188657 0.365806 -0.349524 0.194516 \n", | |
| "precipitate 0.077672 0.176044 -0.085269 -0.121363 \n", | |
| "propranolol_hcl 0.499701 -0.210794 0.384330 0.271781 \n", | |
| "multivitamin/multimineral 0.620430 -0.242618 0.506193 0.412268 \n", | |
| "break 0.291206 -0.034012 0.350973 -0.255148 \n", | |
| "stability_failure 0.172792 0.359987 -0.277620 -0.480863 \n", | |
| "preservative 0.584663 0.121469 0.303326 0.085117 \n", | |
| "stainless_steel 0.963554 0.276743 0.540535 -0.150424 \n", | |
| "do_not 0.392217 0.016973 0.350163 0.054419 \n", | |
| "good_manufacturing_practices 0.272429 -0.044322 0.843428 0.257539 \n", | |
| "calcium 0.694440 0.153814 0.544600 0.299468 \n", | |
| "discoloration 0.570476 -0.397453 -0.115908 0.058452 \n", | |
| "approve_nda/anda 0.008002 0.324189 -0.094483 0.583553 \n", | |
| "substance 0.566097 -0.366434 -0.060759 0.059307 \n", | |
| "insufficient_datum 0.068721 0.452180 -0.211653 -0.494282 \n", | |
| "dr 0.650217 -0.068841 0.551029 0.457740 \n", | |
| "salicylic_acid 0.050387 -0.023391 0.131366 0.421586 \n", | |
| "stability_time_point -0.302260 -0.045905 0.337051 0.115155 \n", | |
| "hence 0.809901 -0.301491 -0.028232 0.252878 \n", | |
| "subpotent_single_ingredient 0.038740 -0.716861 0.435336 0.349279 \n", | |
| "lozenge 0.297437 -0.325696 0.115757 0.026851 \n", | |
| "previous_lot_there 0.107292 0.420130 -0.265406 -0.533381 \n", | |
| "determine_that_other -0.008785 0.432847 -0.190846 -0.468886 \n", | |
| "burkholderia_cepacia 1.087692 -0.327379 0.203308 0.325728 \n", | |
| "market_without 0.055534 0.111120 -0.203184 0.478466 \n", | |
| "quality_review 0.124640 0.316804 -0.235937 -0.510923 \n", | |
| "tadalafil 0.320275 -0.436805 0.639454 0.280085 \n", | |
| "manufacturing_firm 1.041262 -0.871369 0.395397 -0.129529 \n", | |
| "\n", | |
| " 95 96 97 98 99 \n", | |
| "of -0.228435 -0.041155 -0.158670 0.077506 -0.346080 \n", | |
| "be -0.001053 -0.013274 0.230379 -0.141617 -0.203462 \n", | |
| "the 0.241228 -0.147339 -0.153858 0.165506 -0.316384 \n", | |
| "sterility -0.191247 0.163880 0.044130 0.070255 -1.068096 \n", | |
| "assurance -0.296976 -0.091350 0.045781 0.393641 -0.406683 \n", | |
| "product -0.198794 0.290512 0.423234 -0.167009 -0.739767 \n", | |
| "lack -0.059666 -0.216124 -0.388248 0.065185 -0.543164 \n", | |
| "and -0.102934 0.405758 0.062987 -0.083985 -0.631277 \n", | |
| "in 0.187467 0.664412 0.068200 -0.363377 -0.660456 \n", | |
| "to 0.104488 0.230591 -0.263950 -0.015821 -0.146170 \n", | |
| "with 0.153057 0.254042 -0.105156 -0.114450 0.147127 \n", | |
| "a -0.039583 0.217508 0.132062 -0.141048 0.212403 \n", | |
| "penicillin -0.213078 0.071543 0.367104 -0.180235 -0.564265 \n", | |
| "all -0.016561 -0.083103 -0.058704 0.028399 -0.643742 \n", | |
| "recall 0.181283 0.224981 0.670257 0.409089 -0.226304 \n", | |
| "tablet 0.216297 0.002845 0.259346 -0.213895 -0.033015 \n", | |
| "repackaged 0.098583 -0.406318 0.367639 -0.038518 -0.390859 \n", | |
| "lot 0.156697 0.123817 -0.069584 -0.631081 -0.503107 \n", | |
| "pedigree 0.331809 -0.893086 0.014228 -0.050464 -0.126028 \n", | |
| "presence -0.535924 0.232693 -0.142808 0.555902 -0.291969 \n", | |
| "due -0.246100 0.389896 -0.395062 0.338876 -0.196595 \n", | |
| "mg_ndc 0.625140 -0.849242 0.208186 0.082466 0.105004 \n", | |
| "mg 0.731755 -0.635973 0.035107 -0.005107 0.035497 \n", | |
| "cross_contamination 0.127917 -0.376043 0.157067 0.389089 0.247304 \n", | |
| "recall_because_they 0.377424 0.086698 0.022968 -0.190156 -0.776980 \n", | |
| "may 0.096305 0.180668 0.100313 0.003281 0.028236 \n", | |
| "quality -0.848754 0.629272 -0.718522 1.154461 -1.393663 \n", | |
| "facility 0.067188 -0.314685 -0.125226 -0.161432 -0.733695 \n", | |
| "02/12/15 -0.528969 -0.073393 0.023062 0.240035 -0.667557 \n", | |
| "distribute_between_01/05/12 0.202878 0.032444 0.106908 0.141060 -0.854404 \n", | |
| "... ... ... ... ... ... \n", | |
| "exp_6/6/2014 0.702510 -0.406129 0.038623 -0.038386 -0.328455 \n", | |
| "exp_6/25/2014 0.717060 -0.492291 -0.359080 0.233809 -0.572923 \n", | |
| "stability_datum_do_not 0.359418 -0.437779 0.079897 0.159291 -0.514144 \n", | |
| "precipitate 0.945627 -0.328054 -0.555913 -0.043336 -0.336172 \n", | |
| "propranolol_hcl 0.706757 -0.971606 -0.094029 0.472163 -0.350248 \n", | |
| "multivitamin/multimineral 0.755777 -0.950551 0.286023 0.243958 -0.134040 \n", | |
| "break -0.001081 -1.175425 -0.038030 0.217955 -0.392921 \n", | |
| "stability_failure 0.743065 -0.382206 -0.209747 0.431726 -0.799726 \n", | |
| "preservative -0.005443 -0.402300 0.329036 -0.382790 -0.156805 \n", | |
| "stainless_steel 0.124394 -0.597058 0.036565 0.143074 -0.331011 \n", | |
| "do_not 0.383941 0.063834 0.500655 -0.475887 -0.417157 \n", | |
| "good_manufacturing_practices -0.003327 0.267934 0.462099 -0.688051 -0.695076 \n", | |
| "calcium 0.412340 -1.166201 -0.020409 0.199746 -0.273912 \n", | |
| "discoloration 0.682327 -0.359661 0.497713 -0.077592 -0.187794 \n", | |
| "approve_nda/anda 0.110497 0.193130 -0.156360 0.187089 -0.964792 \n", | |
| "substance 0.049056 0.058596 0.254278 -0.088240 -0.340175 \n", | |
| "insufficient_datum 0.595109 -0.310056 0.064565 0.471530 -0.737062 \n", | |
| "dr 0.564364 -0.939611 0.338181 0.279912 -0.783463 \n", | |
| "salicylic_acid 0.224686 -0.071084 0.177473 0.115413 -0.940842 \n", | |
| "stability_time_point 0.355597 -0.358764 -0.141571 0.444522 -1.462083 \n", | |
| "hence 0.051660 -0.016800 -0.362617 0.699495 -0.971199 \n", | |
| "subpotent_single_ingredient 0.618122 -0.182675 0.425007 -0.030217 -0.608149 \n", | |
| "lozenge 0.231624 -0.345866 0.419189 -0.216863 -0.675952 \n", | |
| "previous_lot_there 0.699781 -0.331206 -0.059230 0.433114 -0.782535 \n", | |
| "determine_that_other 0.472048 -0.324222 0.156423 0.405998 -0.654671 \n", | |
| "burkholderia_cepacia -0.273550 0.297474 0.830083 -0.312336 0.019949 \n", | |
| "market_without -0.062840 0.127311 -0.001967 0.144946 -0.978934 \n", | |
| "quality_review 0.877442 -0.289462 -0.270052 0.417941 -0.803751 \n", | |
| "tadalafil -0.091469 -0.390900 0.375969 -0.062292 -0.337352 \n", | |
| "manufacturing_firm 0.370408 -0.257274 0.303240 -0.300379 -0.106373 \n", | |
| "\n", | |
| "[422 rows x 100 columns]" | |
| ] | |
| }, | |
| "execution_count": 62, | |
| "metadata": {}, | |
| "output_type": "execute_result" | |
| } | |
| ], | |
| "source": [ | |
| "# build a list of the terms, integer indices,\n", | |
| "# and term counts from the food2vec model vocabulary\n", | |
| "ordered_vocab = [(term, voc.index, voc.count)\n", | |
| " for term, voc in recall2vec.wv.vocab.items()]\n", | |
| "\n", | |
| "# sort by the term counts, so the most common terms appear first\n", | |
| "ordered_vocab = sorted(ordered_vocab, key=lambda term_index: -term_index[2])\n", | |
| "\n", | |
| "# unzip the terms, integer indices, and counts into separate lists\n", | |
| "ordered_terms, term_indices, term_counts = zip(*ordered_vocab)\n", | |
| "\n", | |
| "# create a DataFrame with the food2vec vectors as data,\n", | |
| "# and the terms as row labels\n", | |
| "word_vectors = pd.DataFrame(recall2vec.wv.syn0[term_indices, :],\n", | |
| " index=ordered_terms)\n", | |
| "\n", | |
| "word_vectors" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "### So... what can we do with all these numbers?\n", | |
| "The first thing we can use them for is to simply look up related words and phrases for a given term of interest." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 63, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "def get_related_terms(token, topn=5):\n", | |
| " \"\"\"\n", | |
| " look up the topn most similar terms to token\n", | |
| " and print them as a formatted list\n", | |
| " \"\"\"\n", | |
| "\n", | |
| " for word, similarity in recall2vec.most_similar(positive=[token], topn=topn):\n", | |
| "\n", | |
| " print('{:20} {}'.format(word, round(similarity, 3)))" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "### Issues with Syringe?" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 65, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "support_expiry_potential_loss 0.68\n", | |
| "store 0.636\n", | |
| "drug_package 0.626\n", | |
| "defective_delivery_system 0.516\n", | |
| "potency 0.492\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "get_related_terms('syringe')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "### What about tablets?" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 66, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "package_insert 0.467\n", | |
| "strength 0.432\n", | |
| "bottle 0.431\n", | |
| "fail_stability 0.399\n", | |
| "contaminate 0.399\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "get_related_terms('tablets')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "### Since we are mining for Drug Recalls, what are the top factors?" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 67, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "initiate 0.427\n", | |
| "poor_sterile 0.405\n", | |
| "production_practice_result 0.404\n", | |
| "due 0.397\n", | |
| "quality_control_procedure 0.355\n", | |
| "by 0.353\n", | |
| "concerned_test_result_obtain 0.345\n", | |
| "05/21/2012 0.341\n", | |
| "laboratory 0.341\n", | |
| "fda 0.341\n", | |
| "contain_undeclared 0.341\n", | |
| "observation_associate 0.34\n", | |
| "tadalafil 0.338\n", | |
| "reliable 0.336\n", | |
| "unapproved_drug 0.33\n", | |
| "inc. 0.326\n", | |
| "laboratory_result 0.325\n", | |
| "be 0.325\n", | |
| "not_expire_due 0.322\n", | |
| "manufacturer 0.317\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "get_related_terms('recall', topn=20)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "## Word algebra!\n", | |
| "No self-respecting word2vec demo would be complete without a healthy dose of *word algebra*, also known as *analogy completion*.\n", | |
| "\n", | |
| "The core idea is that once words are represented as numerical vectors, you can do math with them. The mathematical procedure goes like this:\n", | |
| "1. Provide a set of words or phrases that you'd like to add or subtract.\n", | |
| "1. Look up the vectors that represent those terms in the word vector model.\n", | |
| "1. Add and subtract those vectors to produce a new, combined vector.\n", | |
| "1. Look up the most similar vector(s) to this new, combined vector via cosine similarity.\n", | |
| "1. Return the word(s) associated with the similar vector(s).\n", | |
| "\n", | |
| "But more generally, you can think of the vectors that represent each word as encoding some information about the *meaning* or *concepts* of the word. What happens when you ask the model to combine the meaning and concepts of words in new ways? Let's see." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 68, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "def word_algebra(add=[], subtract=[], topn=1):\n", | |
| " \"\"\"\n", | |
| " combine the vectors associated with the words provided\n", | |
| " in add= and subtract=, look up the topn most similar\n", | |
| " terms to the combined vector, and print the result(s)\n", | |
| " \"\"\"\n", | |
| " answers = recall2vec.most_similar(positive=add, negative=subtract, topn=topn)\n", | |
| " \n", | |
| " for term, similarity in answers:\n", | |
| " print(term)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "### ?" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 109, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "stainless_steel\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "word_algebra(add=['presence', 'calcium'])" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "### ?" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 100, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "support_expiry_recent\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "word_algebra(add=['sterility', 'syringe'], subtract=['label'])" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "### ?" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 108, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "sibutramine\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "word_algebra(add=['fda', 'product'], subtract=['sterile'])" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "## Word Vector Visualization with t-SNE" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "[t-Distributed Stochastic Neighbor Embedding](https://lvdmaaten.github.io/publications/papers/JMLR_2008.pdf), or *t-SNE* for short, is a dimensionality reduction technique to assist with visualizing high-dimensional datasets. It attempts to map high-dimensional data onto a low two- or three-dimensional representation such that the relative distances between points are preserved as closely as possible in both high-dimensional and low-dimensional space.\n", | |
| "\n", | |
| "scikit-learn provides a convenient implementation of the t-SNE algorithm with its [TSNE](http://scikit-learn.org/stable/modules/generated/sklearn.manifold.TSNE.html) class." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 91, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "from sklearn.manifold import TSNE" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Our input for t-SNE will be the DataFrame of word vectors we created before. Let's first:\n", | |
| "1. Drop stopwords — it's probably not too interesting to visualize *the*, *of*, *or*, and so on\n", | |
| "1. Take only the 5,000 most frequent terms in the vocabulary — no need to visualize all ~50,000 terms right now." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 92, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "tsne_input = word_vectors.drop(spacy.en.stop_words.STOP_WORDS, errors='ignore')\n", | |
| "tsne_input = tsne_input.head(1000)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 93, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "data": { | |
| "text/html": [ | |
| "<div>\n", | |
| "<table border=\"1\" class=\"dataframe\">\n", | |
| " <thead>\n", | |
| " <tr style=\"text-align: right;\">\n", | |
| " <th></th>\n", | |
| " <th>0</th>\n", | |
| " <th>1</th>\n", | |
| " <th>2</th>\n", | |
| " <th>3</th>\n", | |
| " <th>4</th>\n", | |
| " <th>5</th>\n", | |
| " <th>6</th>\n", | |
| " <th>7</th>\n", | |
| " <th>8</th>\n", | |
| " <th>9</th>\n", | |
| " <th>...</th>\n", | |
| " <th>90</th>\n", | |
| " <th>91</th>\n", | |
| " <th>92</th>\n", | |
| " <th>93</th>\n", | |
| " <th>94</th>\n", | |
| " <th>95</th>\n", | |
| " <th>96</th>\n", | |
| " <th>97</th>\n", | |
| " <th>98</th>\n", | |
| " <th>99</th>\n", | |
| " </tr>\n", | |
| " </thead>\n", | |
| " <tbody>\n", | |
| " <tr>\n", | |
| " <th>sterility</th>\n", | |
| " <td>0.303488</td>\n", | |
| " <td>-0.228153</td>\n", | |
| " <td>-0.219394</td>\n", | |
| " <td>-0.302188</td>\n", | |
| " <td>-0.070613</td>\n", | |
| " <td>0.450770</td>\n", | |
| " <td>-0.125470</td>\n", | |
| " <td>-0.174656</td>\n", | |
| " <td>0.334442</td>\n", | |
| " <td>0.810358</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.372313</td>\n", | |
| " <td>0.377984</td>\n", | |
| " <td>-0.309888</td>\n", | |
| " <td>-0.746572</td>\n", | |
| " <td>-0.342844</td>\n", | |
| " <td>-0.191247</td>\n", | |
| " <td>0.163880</td>\n", | |
| " <td>0.044130</td>\n", | |
| " <td>0.070255</td>\n", | |
| " <td>-1.068096</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>assurance</th>\n", | |
| " <td>0.240375</td>\n", | |
| " <td>-0.008034</td>\n", | |
| " <td>-0.594212</td>\n", | |
| " <td>0.349680</td>\n", | |
| " <td>-0.603484</td>\n", | |
| " <td>0.530704</td>\n", | |
| " <td>0.029388</td>\n", | |
| " <td>0.626848</td>\n", | |
| " <td>0.348065</td>\n", | |
| " <td>0.077353</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.388538</td>\n", | |
| " <td>0.002734</td>\n", | |
| " <td>-0.239271</td>\n", | |
| " <td>-0.082077</td>\n", | |
| " <td>-0.047349</td>\n", | |
| " <td>-0.296976</td>\n", | |
| " <td>-0.091350</td>\n", | |
| " <td>0.045781</td>\n", | |
| " <td>0.393641</td>\n", | |
| " <td>-0.406683</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>product</th>\n", | |
| " <td>0.167806</td>\n", | |
| " <td>-0.021859</td>\n", | |
| " <td>0.185118</td>\n", | |
| " <td>-0.030679</td>\n", | |
| " <td>-0.188094</td>\n", | |
| " <td>0.140964</td>\n", | |
| " <td>-0.239155</td>\n", | |
| " <td>0.544782</td>\n", | |
| " <td>0.352739</td>\n", | |
| " <td>0.016653</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.025098</td>\n", | |
| " <td>-0.186757</td>\n", | |
| " <td>-0.083697</td>\n", | |
| " <td>-0.093634</td>\n", | |
| " <td>-0.106546</td>\n", | |
| " <td>-0.198794</td>\n", | |
| " <td>0.290512</td>\n", | |
| " <td>0.423234</td>\n", | |
| " <td>-0.167009</td>\n", | |
| " <td>-0.739767</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>lack</th>\n", | |
| " <td>0.666243</td>\n", | |
| " <td>-0.235590</td>\n", | |
| " <td>-0.383695</td>\n", | |
| " <td>0.450461</td>\n", | |
| " <td>-0.533766</td>\n", | |
| " <td>0.610448</td>\n", | |
| " <td>-0.394387</td>\n", | |
| " <td>0.135181</td>\n", | |
| " <td>-0.221998</td>\n", | |
| " <td>0.284320</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.096456</td>\n", | |
| " <td>0.144373</td>\n", | |
| " <td>0.413397</td>\n", | |
| " <td>-0.051289</td>\n", | |
| " <td>-0.165963</td>\n", | |
| " <td>-0.059666</td>\n", | |
| " <td>-0.216124</td>\n", | |
| " <td>-0.388248</td>\n", | |
| " <td>0.065185</td>\n", | |
| " <td>-0.543164</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>penicillin</th>\n", | |
| " <td>0.331125</td>\n", | |
| " <td>0.195782</td>\n", | |
| " <td>-0.216822</td>\n", | |
| " <td>-0.540945</td>\n", | |
| " <td>-0.070391</td>\n", | |
| " <td>-0.168526</td>\n", | |
| " <td>0.119359</td>\n", | |
| " <td>0.104435</td>\n", | |
| " <td>-0.079008</td>\n", | |
| " <td>0.072145</td>\n", | |
| " <td>...</td>\n", | |
| " <td>0.097845</td>\n", | |
| " <td>0.443677</td>\n", | |
| " <td>-0.943778</td>\n", | |
| " <td>-0.739152</td>\n", | |
| " <td>0.490247</td>\n", | |
| " <td>-0.213078</td>\n", | |
| " <td>0.071543</td>\n", | |
| " <td>0.367104</td>\n", | |
| " <td>-0.180235</td>\n", | |
| " <td>-0.564265</td>\n", | |
| " </tr>\n", | |
| " </tbody>\n", | |
| "</table>\n", | |
| "<p>5 rows × 100 columns</p>\n", | |
| "</div>" | |
| ], | |
| "text/plain": [ | |
| " 0 1 2 3 4 5 \\\n", | |
| "sterility 0.303488 -0.228153 -0.219394 -0.302188 -0.070613 0.450770 \n", | |
| "assurance 0.240375 -0.008034 -0.594212 0.349680 -0.603484 0.530704 \n", | |
| "product 0.167806 -0.021859 0.185118 -0.030679 -0.188094 0.140964 \n", | |
| "lack 0.666243 -0.235590 -0.383695 0.450461 -0.533766 0.610448 \n", | |
| "penicillin 0.331125 0.195782 -0.216822 -0.540945 -0.070391 -0.168526 \n", | |
| "\n", | |
| " 6 7 8 9 ... 90 \\\n", | |
| "sterility -0.125470 -0.174656 0.334442 0.810358 ... 0.372313 \n", | |
| "assurance 0.029388 0.626848 0.348065 0.077353 ... 0.388538 \n", | |
| "product -0.239155 0.544782 0.352739 0.016653 ... 0.025098 \n", | |
| "lack -0.394387 0.135181 -0.221998 0.284320 ... 0.096456 \n", | |
| "penicillin 0.119359 0.104435 -0.079008 0.072145 ... 0.097845 \n", | |
| "\n", | |
| " 91 92 93 94 95 96 \\\n", | |
| "sterility 0.377984 -0.309888 -0.746572 -0.342844 -0.191247 0.163880 \n", | |
| "assurance 0.002734 -0.239271 -0.082077 -0.047349 -0.296976 -0.091350 \n", | |
| "product -0.186757 -0.083697 -0.093634 -0.106546 -0.198794 0.290512 \n", | |
| "lack 0.144373 0.413397 -0.051289 -0.165963 -0.059666 -0.216124 \n", | |
| "penicillin 0.443677 -0.943778 -0.739152 0.490247 -0.213078 0.071543 \n", | |
| "\n", | |
| " 97 98 99 \n", | |
| "sterility 0.044130 0.070255 -1.068096 \n", | |
| "assurance 0.045781 0.393641 -0.406683 \n", | |
| "product 0.423234 -0.167009 -0.739767 \n", | |
| "lack -0.388248 0.065185 -0.543164 \n", | |
| "penicillin 0.367104 -0.180235 -0.564265 \n", | |
| "\n", | |
| "[5 rows x 100 columns]" | |
| ] | |
| }, | |
| "execution_count": 93, | |
| "metadata": {}, | |
| "output_type": "execute_result" | |
| } | |
| ], | |
| "source": [ | |
| "tsne_input.head()" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 94, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "tsne_filepath = os.path.join(intermediate_directory,\n", | |
| " 'tsne_model')\n", | |
| "\n", | |
| "tsne_vectors_filepath = os.path.join(intermediate_directory,\n", | |
| " 'tsne_vectors.npy')" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 95, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "Wall time: 64 ms\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "%%time\n", | |
| "\n", | |
| "if 0 == 1:\n", | |
| " \n", | |
| " tsne = TSNE()\n", | |
| " tsne_vectors = tsne.fit_transform(tsne_input.values)\n", | |
| " \n", | |
| " with open(tsne_filepath, 'wb') as f:\n", | |
| " pickle.dump(tsne, f)\n", | |
| "\n", | |
| " pd.np.save(tsne_vectors_filepath, tsne_vectors)\n", | |
| " \n", | |
| "with open(tsne_filepath, \"rb\") as f:\n", | |
| " tsne = pickle.load(f)\n", | |
| " \n", | |
| "tsne_vectors = pd.np.load(tsne_vectors_filepath)\n", | |
| "\n", | |
| "tsne_vectors = pd.DataFrame(tsne_vectors,\n", | |
| " index=pd.Index(tsne_input.index),\n", | |
| " columns=['x_coord', 'y_coord'])" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Now we have a two-dimensional representation of our data! Let's take a look." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 96, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "data": { | |
| "text/html": [ | |
| "<div>\n", | |
| "<table border=\"1\" class=\"dataframe\">\n", | |
| " <thead>\n", | |
| " <tr style=\"text-align: right;\">\n", | |
| " <th></th>\n", | |
| " <th>x_coord</th>\n", | |
| " <th>y_coord</th>\n", | |
| " </tr>\n", | |
| " </thead>\n", | |
| " <tbody>\n", | |
| " <tr>\n", | |
| " <th>sterility</th>\n", | |
| " <td>-8.427881</td>\n", | |
| " <td>15.936145</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>assurance</th>\n", | |
| " <td>7.898233</td>\n", | |
| " <td>-1.578057</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>product</th>\n", | |
| " <td>-20.207019</td>\n", | |
| " <td>2.562714</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>lack</th>\n", | |
| " <td>-26.316070</td>\n", | |
| " <td>3.674049</td>\n", | |
| " </tr>\n", | |
| " <tr>\n", | |
| " <th>penicillin</th>\n", | |
| " <td>47.524378</td>\n", | |
| " <td>11.912826</td>\n", | |
| " </tr>\n", | |
| " </tbody>\n", | |
| "</table>\n", | |
| "</div>" | |
| ], | |
| "text/plain": [ | |
| " x_coord y_coord\n", | |
| "sterility -8.427881 15.936145\n", | |
| "assurance 7.898233 -1.578057\n", | |
| "product -20.207019 2.562714\n", | |
| "lack -26.316070 3.674049\n", | |
| "penicillin 47.524378 11.912826" | |
| ] | |
| }, | |
| "execution_count": 96, | |
| "metadata": {}, | |
| "output_type": "execute_result" | |
| } | |
| ], | |
| "source": [ | |
| "tsne_vectors.head()" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 97, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "tsne_vectors['word'] = tsne_vectors.index" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "### Plotting with Bokeh" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 98, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stderr", | |
| "output_type": "stream", | |
| "text": [ | |
| "C:\\Users\\aashis_tiwari\\AppData\\Local\\Continuum\\Anaconda3\\envs\\tensorflow\\lib\\site-packages\\bokeh\\core\\json_encoder.py:52: DeprecationWarning: parsing timezone aware datetimes is deprecated; this will raise an error in the future\n", | |
| " NP_EPOCH = np.datetime64('1970-01-01T00:00:00Z')\n" | |
| ] | |
| }, | |
| { | |
| "data": { | |
| "text/html": [ | |
| "\n", | |
| " <div class=\"bk-root\">\n", | |
| " <a href=\"http://bokeh.pydata.org\" target=\"_blank\" class=\"bk-logo bk-logo-small bk-logo-notebook\"></a>\n", | |
| " <span id=\"79b89acc-ad85-4e61-a6d7-d7fee4eefc13\">Loading BokehJS ...</span>\n", | |
| " </div>" | |
| ] | |
| }, | |
| "metadata": {}, | |
| "output_type": "display_data" | |
| }, | |
| { | |
| "data": { | |
| "application/javascript": [ | |
| "\n", | |
| "(function(global) {\n", | |
| " function now() {\n", | |
| " return new Date();\n", | |
| " }\n", | |
| "\n", | |
| " var force = true;\n", | |
| "\n", | |
| " if (typeof (window._bokeh_onload_callbacks) === \"undefined\" || force === true) {\n", | |
| " window._bokeh_onload_callbacks = [];\n", | |
| " window._bokeh_is_loading = undefined;\n", | |
| " }\n", | |
| "\n", | |
| "\n", | |
| " \n", | |
| " if (typeof (window._bokeh_timeout) === \"undefined\" || force === true) {\n", | |
| " window._bokeh_timeout = Date.now() + 5000;\n", | |
| " window._bokeh_failed_load = false;\n", | |
| " }\n", | |
| "\n", | |
| " var NB_LOAD_WARNING = {'data': {'text/html':\n", | |
| " \"<div style='background-color: #fdd'>\\n\"+\n", | |
| " \"<p>\\n\"+\n", | |
| " \"BokehJS does not appear to have successfully loaded. If loading BokehJS from CDN, this \\n\"+\n", | |
| " \"may be due to a slow or bad network connection. Possible fixes:\\n\"+\n", | |
| " \"</p>\\n\"+\n", | |
| " \"<ul>\\n\"+\n", | |
| " \"<li>re-rerun `output_notebook()` to attempt to load from CDN again, or</li>\\n\"+\n", | |
| " \"<li>use INLINE resources instead, as so:</li>\\n\"+\n", | |
| " \"</ul>\\n\"+\n", | |
| " \"<code>\\n\"+\n", | |
| " \"from bokeh.resources import INLINE\\n\"+\n", | |
| " \"output_notebook(resources=INLINE)\\n\"+\n", | |
| " \"</code>\\n\"+\n", | |
| " \"</div>\"}};\n", | |
| "\n", | |
| " function display_loaded() {\n", | |
| " if (window.Bokeh !== undefined) {\n", | |
| " document.getElementById(\"79b89acc-ad85-4e61-a6d7-d7fee4eefc13\").textContent = \"BokehJS successfully loaded.\";\n", | |
| " } else if (Date.now() < window._bokeh_timeout) {\n", | |
| " setTimeout(display_loaded, 100)\n", | |
| " }\n", | |
| " }\n", | |
| "\n", | |
| " function run_callbacks() {\n", | |
| " window._bokeh_onload_callbacks.forEach(function(callback) { callback() });\n", | |
| " delete window._bokeh_onload_callbacks\n", | |
| " console.info(\"Bokeh: all callbacks have finished\");\n", | |
| " }\n", | |
| "\n", | |
| " function load_libs(js_urls, callback) {\n", | |
| " window._bokeh_onload_callbacks.push(callback);\n", | |
| " if (window._bokeh_is_loading > 0) {\n", | |
| " console.log(\"Bokeh: BokehJS is being loaded, scheduling callback at\", now());\n", | |
| " return null;\n", | |
| " }\n", | |
| " if (js_urls == null || js_urls.length === 0) {\n", | |
| " run_callbacks();\n", | |
| " return null;\n", | |
| " }\n", | |
| " console.log(\"Bokeh: BokehJS not loaded, scheduling load and callback at\", now());\n", | |
| " window._bokeh_is_loading = js_urls.length;\n", | |
| " for (var i = 0; i < js_urls.length; i++) {\n", | |
| " var url = js_urls[i];\n", | |
| " var s = document.createElement('script');\n", | |
| " s.src = url;\n", | |
| " s.async = false;\n", | |
| " s.onreadystatechange = s.onload = function() {\n", | |
| " window._bokeh_is_loading--;\n", | |
| " if (window._bokeh_is_loading === 0) {\n", | |
| " console.log(\"Bokeh: all BokehJS libraries loaded\");\n", | |
| " run_callbacks()\n", | |
| " }\n", | |
| " };\n", | |
| " s.onerror = function() {\n", | |
| " console.warn(\"failed to load library \" + url);\n", | |
| " };\n", | |
| " console.log(\"Bokeh: injecting script tag for BokehJS library: \", url);\n", | |
| " document.getElementsByTagName(\"head\")[0].appendChild(s);\n", | |
| " }\n", | |
| " };var element = document.getElementById(\"79b89acc-ad85-4e61-a6d7-d7fee4eefc13\");\n", | |
| " if (element == null) {\n", | |
| " console.log(\"Bokeh: ERROR: autoload.js configured with elementid '79b89acc-ad85-4e61-a6d7-d7fee4eefc13' but no matching script tag was found. \")\n", | |
| " return false;\n", | |
| " }\n", | |
| "\n", | |
| " var js_urls = [\"https://cdn.pydata.org/bokeh/release/bokeh-0.12.4.min.js\", \"https://cdn.pydata.org/bokeh/release/bokeh-widgets-0.12.4.min.js\"];\n", | |
| "\n", | |
| " var inline_js = [\n", | |
| " function(Bokeh) {\n", | |
| " Bokeh.set_log_level(\"info\");\n", | |
| " },\n", | |
| " \n", | |
| " function(Bokeh) {\n", | |
| " \n", | |
| " document.getElementById(\"79b89acc-ad85-4e61-a6d7-d7fee4eefc13\").textContent = \"BokehJS is loading...\";\n", | |
| " },\n", | |
| " function(Bokeh) {\n", | |
| " console.log(\"Bokeh: injecting CSS: https://cdn.pydata.org/bokeh/release/bokeh-0.12.4.min.css\");\n", | |
| " Bokeh.embed.inject_css(\"https://cdn.pydata.org/bokeh/release/bokeh-0.12.4.min.css\");\n", | |
| " console.log(\"Bokeh: injecting CSS: https://cdn.pydata.org/bokeh/release/bokeh-widgets-0.12.4.min.css\");\n", | |
| " Bokeh.embed.inject_css(\"https://cdn.pydata.org/bokeh/release/bokeh-widgets-0.12.4.min.css\");\n", | |
| " }\n", | |
| " ];\n", | |
| "\n", | |
| " function run_inline_js() {\n", | |
| " \n", | |
| " if ((window.Bokeh !== undefined) || (force === true)) {\n", | |
| " for (var i = 0; i < inline_js.length; i++) {\n", | |
| " inline_js[i](window.Bokeh);\n", | |
| " }if (force === true) {\n", | |
| " display_loaded();\n", | |
| " }} else if (Date.now() < window._bokeh_timeout) {\n", | |
| " setTimeout(run_inline_js, 100);\n", | |
| " } else if (!window._bokeh_failed_load) {\n", | |
| " console.log(\"Bokeh: BokehJS failed to load within specified timeout.\");\n", | |
| " window._bokeh_failed_load = true;\n", | |
| " } else if (force !== true) {\n", | |
| " var cell = $(document.getElementById(\"79b89acc-ad85-4e61-a6d7-d7fee4eefc13\")).parents('.cell').data().cell;\n", | |
| " cell.output_area.append_execute_result(NB_LOAD_WARNING)\n", | |
| " }\n", | |
| "\n", | |
| " }\n", | |
| "\n", | |
| " if (window._bokeh_is_loading === 0) {\n", | |
| " console.log(\"Bokeh: BokehJS loaded, going straight to plotting\");\n", | |
| " run_inline_js();\n", | |
| " } else {\n", | |
| " load_libs(js_urls, function() {\n", | |
| " console.log(\"Bokeh: BokehJS plotting callback run at\", now());\n", | |
| " run_inline_js();\n", | |
| " });\n", | |
| " }\n", | |
| "}(this));" | |
| ] | |
| }, | |
| "metadata": {}, | |
| "output_type": "display_data" | |
| } | |
| ], | |
| "source": [ | |
| "from bokeh.plotting import figure, show, output_notebook\n", | |
| "from bokeh.models import HoverTool, ColumnDataSource, value\n", | |
| "\n", | |
| "output_notebook()" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 99, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "data": { | |
| "text/html": [ | |
| "\n", | |
| "\n", | |
| " <div class=\"bk-root\">\n", | |
| " <div class=\"bk-plotdiv\" id=\"1ffba516-d86c-45bc-9d9c-f02e7c30c12b\"></div>\n", | |
| " </div>\n", | |
| "<script type=\"text/javascript\">\n", | |
| " \n", | |
| " (function(global) {\n", | |
| " function now() {\n", | |
| " return new Date();\n", | |
| " }\n", | |
| " \n", | |
| " var force = false;\n", | |
| " \n", | |
| " if (typeof (window._bokeh_onload_callbacks) === \"undefined\" || force === true) {\n", | |
| " window._bokeh_onload_callbacks = [];\n", | |
| " window._bokeh_is_loading = undefined;\n", | |
| " }\n", | |
| " \n", | |
| " \n", | |
| " \n", | |
| " if (typeof (window._bokeh_timeout) === \"undefined\" || force === true) {\n", | |
| " window._bokeh_timeout = Date.now() + 0;\n", | |
| " window._bokeh_failed_load = false;\n", | |
| " }\n", | |
| " \n", | |
| " var NB_LOAD_WARNING = {'data': {'text/html':\n", | |
| " \"<div style='background-color: #fdd'>\\n\"+\n", | |
| " \"<p>\\n\"+\n", | |
| " \"BokehJS does not appear to have successfully loaded. If loading BokehJS from CDN, this \\n\"+\n", | |
| " \"may be due to a slow or bad network connection. Possible fixes:\\n\"+\n", | |
| " \"</p>\\n\"+\n", | |
| " \"<ul>\\n\"+\n", | |
| " \"<li>re-rerun `output_notebook()` to attempt to load from CDN again, or</li>\\n\"+\n", | |
| " \"<li>use INLINE resources instead, as so:</li>\\n\"+\n", | |
| " \"</ul>\\n\"+\n", | |
| " \"<code>\\n\"+\n", | |
| " \"from bokeh.resources import INLINE\\n\"+\n", | |
| " \"output_notebook(resources=INLINE)\\n\"+\n", | |
| " \"</code>\\n\"+\n", | |
| " \"</div>\"}};\n", | |
| " \n", | |
| " function display_loaded() {\n", | |
| " if (window.Bokeh !== undefined) {\n", | |
| " document.getElementById(\"1ffba516-d86c-45bc-9d9c-f02e7c30c12b\").textContent = \"BokehJS successfully loaded.\";\n", | |
| " } else if (Date.now() < window._bokeh_timeout) {\n", | |
| " setTimeout(display_loaded, 100)\n", | |
| " }\n", | |
| " }\n", | |
| " \n", | |
| " function run_callbacks() {\n", | |
| " window._bokeh_onload_callbacks.forEach(function(callback) { callback() });\n", | |
| " delete window._bokeh_onload_callbacks\n", | |
| " console.info(\"Bokeh: all callbacks have finished\");\n", | |
| " }\n", | |
| " \n", | |
| " function load_libs(js_urls, callback) {\n", | |
| " window._bokeh_onload_callbacks.push(callback);\n", | |
| " if (window._bokeh_is_loading > 0) {\n", | |
| " console.log(\"Bokeh: BokehJS is being loaded, scheduling callback at\", now());\n", | |
| " return null;\n", | |
| " }\n", | |
| " if (js_urls == null || js_urls.length === 0) {\n", | |
| " run_callbacks();\n", | |
| " return null;\n", | |
| " }\n", | |
| " console.log(\"Bokeh: BokehJS not loaded, scheduling load and callback at\", now());\n", | |
| " window._bokeh_is_loading = js_urls.length;\n", | |
| " for (var i = 0; i < js_urls.length; i++) {\n", | |
| " var url = js_urls[i];\n", | |
| " var s = document.createElement('script');\n", | |
| " s.src = url;\n", | |
| " s.async = false;\n", | |
| " s.onreadystatechange = s.onload = function() {\n", | |
| " window._bokeh_is_loading--;\n", | |
| " if (window._bokeh_is_loading === 0) {\n", | |
| " console.log(\"Bokeh: all BokehJS libraries loaded\");\n", | |
| " run_callbacks()\n", | |
| " }\n", | |
| " };\n", | |
| " s.onerror = function() {\n", | |
| " console.warn(\"failed to load library \" + url);\n", | |
| " };\n", | |
| " console.log(\"Bokeh: injecting script tag for BokehJS library: \", url);\n", | |
| " document.getElementsByTagName(\"head\")[0].appendChild(s);\n", | |
| " }\n", | |
| " };var element = document.getElementById(\"1ffba516-d86c-45bc-9d9c-f02e7c30c12b\");\n", | |
| " if (element == null) {\n", | |
| " console.log(\"Bokeh: ERROR: autoload.js configured with elementid '1ffba516-d86c-45bc-9d9c-f02e7c30c12b' but no matching script tag was found. \")\n", | |
| " return false;\n", | |
| " }\n", | |
| " \n", | |
| " var js_urls = [];\n", | |
| " \n", | |
| " var inline_js = [\n", | |
| " function(Bokeh) {\n", | |
| " (function() {\n", | |
| " var fn = function() {\n", | |
| " var docs_json = {\"0da6728f-c9ad-45fd-ad12-9bb446d72c06\":{\"roots\":{\"references\":[{\"attributes\":{\"formatter\":{\"id\":\"2903f9f7-248d-4fd6-aa90-e166da048a2c\",\"type\":\"BasicTickFormatter\"},\"plot\":{\"id\":\"7c81ad3e-90a5-4aae-ac9f-81c66a17c864\",\"subtype\":\"Figure\",\"type\":\"Plot\"},\"ticker\":{\"id\":\"d95a48af-221a-4b1e-8983-71cd170770ce\",\"type\":\"BasicTicker\"},\"visible\":false},\"id\":\"3c1b569e-ad33-4c72-ab55-a068df0c2459\",\"type\":\"LinearAxis\"},{\"attributes\":{},\"id\":\"6ba3c1ab-c854-49ba-9cc0-331583770b2e\",\"type\":\"ToolEvents\"},{\"attributes\":{\"bottom_units\":\"screen\",\"fill_alpha\":{\"value\":0.5},\"fill_color\":{\"value\":\"lightgrey\"},\"left_units\":\"screen\",\"level\":\"overlay\",\"line_alpha\":{\"value\":1.0},\"line_color\":{\"value\":\"black\"},\"line_dash\":[4,4],\"line_width\":{\"value\":2},\"plot\":null,\"render_mode\":\"css\",\"right_units\":\"screen\",\"top_units\":\"screen\"},\"id\":\"cdf2cfc2-3a5c-4644-bfc0-d0adc36bb556\",\"type\":\"BoxAnnotation\"},{\"attributes\":{\"below\":[{\"id\":\"3c1b569e-ad33-4c72-ab55-a068df0c2459\",\"type\":\"LinearAxis\"}],\"left\":[{\"id\":\"fb5c7de4-3ca4-4cfd-857b-efbf045de68f\",\"type\":\"LinearAxis\"}],\"outline_line_color\":{\"value\":null},\"plot_height\":800,\"plot_width\":800,\"renderers\":[{\"id\":\"3c1b569e-ad33-4c72-ab55-a068df0c2459\",\"type\":\"LinearAxis\"},{\"id\":\"7a83ff8e-44dd-4638-bd02-c02a21f46836\",\"type\":\"Grid\"},{\"id\":\"fb5c7de4-3ca4-4cfd-857b-efbf045de68f\",\"type\":\"LinearAxis\"},{\"id\":\"dc564fe1-78fa-42fb-ba0d-bee1bf0d3225\",\"type\":\"Grid\"},{\"id\":\"cdf2cfc2-3a5c-4644-bfc0-d0adc36bb556\",\"type\":\"BoxAnnotation\"},{\"id\":\"973ffbda-0da6-4ed0-a121-9553c8b6fa2d\",\"type\":\"BoxAnnotation\"},{\"id\":\"c5b7dc8a-59ff-4f57-a1e7-4be678d01e5a\",\"type\":\"GlyphRenderer\"}],\"title\":{\"id\":\"37945968-dfc6-46b7-94a6-2dfdf351bf18\",\"type\":\"Title\"},\"tool_events\":{\"id\":\"6ba3c1ab-c854-49ba-9cc0-331583770b2e\",\"type\":\"ToolEvents\"},\"toolbar\":{\"id\":\"f10872fd-a808-43c7-bf37-2c5bd9589f0a\",\"type\":\"Toolbar\"},\"x_range\":{\"id\":\"0a89d6b5-9c7a-4695-881e-db58ae17bde9\",\"type\":\"DataRange1d\"},\"y_range\":{\"id\":\"27653325-c718-476d-b976-82abab131d6d\",\"type\":\"DataRange1d\"}},\"id\":\"7c81ad3e-90a5-4aae-ac9f-81c66a17c864\",\"subtype\":\"Figure\",\"type\":\"Plot\"},{\"attributes\":{\"callback\":null},\"id\":\"27653325-c718-476d-b976-82abab131d6d\",\"type\":\"DataRange1d\"},{\"attributes\":{\"fill_color\":{\"value\":\"#1f77b4\"},\"size\":{\"units\":\"screen\",\"value\":10},\"x\":{\"field\":\"x_coord\"},\"y\":{\"field\":\"y_coord\"}},\"id\":\"93797eac-3d20-467b-8474-f5c85a86cd6d\",\"type\":\"Circle\"},{\"attributes\":{\"grid_line_color\":{\"value\":null},\"plot\":{\"id\":\"7c81ad3e-90a5-4aae-ac9f-81c66a17c864\",\"subtype\":\"Figure\",\"type\":\"Plot\"},\"ticker\":{\"id\":\"d95a48af-221a-4b1e-8983-71cd170770ce\",\"type\":\"BasicTicker\"}},\"id\":\"7a83ff8e-44dd-4638-bd02-c02a21f46836\",\"type\":\"Grid\"},{\"attributes\":{\"bottom_units\":\"screen\",\"fill_alpha\":{\"value\":0.5},\"fill_color\":{\"value\":\"lightgrey\"},\"left_units\":\"screen\",\"level\":\"overlay\",\"line_alpha\":{\"value\":1.0},\"line_color\":{\"value\":\"black\"},\"line_dash\":[4,4],\"line_width\":{\"value\":2},\"plot\":null,\"render_mode\":\"css\",\"right_units\":\"screen\",\"top_units\":\"screen\"},\"id\":\"973ffbda-0da6-4ed0-a121-9553c8b6fa2d\",\"type\":\"BoxAnnotation\"},{\"attributes\":{\"callback\":null,\"overlay\":{\"id\":\"973ffbda-0da6-4ed0-a121-9553c8b6fa2d\",\"type\":\"BoxAnnotation\"},\"plot\":{\"id\":\"7c81ad3e-90a5-4aae-ac9f-81c66a17c864\",\"subtype\":\"Figure\",\"type\":\"Plot\"},\"renderers\":[{\"id\":\"c5b7dc8a-59ff-4f57-a1e7-4be678d01e5a\",\"type\":\"GlyphRenderer\"}]},\"id\":\"051447f1-c457-4aa6-8316-e0f83b0eff6d\",\"type\":\"BoxSelectTool\"},{\"attributes\":{\"fill_alpha\":{\"value\":0.1},\"fill_color\":{\"value\":\"#1f77b4\"},\"line_alpha\":{\"value\":0.1},\"line_color\":{\"value\":\"#1f77b4\"},\"size\":{\"units\":\"screen\",\"value\":10},\"x\":{\"field\":\"x_coord\"},\"y\":{\"field\":\"y_coord\"}},\"id\":\"9ef5454f-f7b8-453e-9f0f-1a95c4510330\",\"type\":\"Circle\"},{\"attributes\":{\"callback\":null,\"plot\":{\"id\":\"7c81ad3e-90a5-4aae-ac9f-81c66a17c864\",\"subtype\":\"Figure\",\"type\":\"Plot\"},\"tooltips\":\"@word\"},\"id\":\"73a8abce-1f48-42e5-9800-f6e5b7730e28\",\"type\":\"HoverTool\"},{\"attributes\":{\"callback\":null,\"column_names\":[\"x_coord\",\"word\",\"y_coord\",\"index\"],\"data\":{\"index\":[\"sterility\",\"assurance\",\"product\",\"lack\",\"penicillin\",\"recall\",\"tablet\",\"repackaged\",\"lot\",\"pedigree\",\"presence\",\"mg_ndc\",\"mg\",\"cross_contamination\",\"recall_because_they\",\"quality\",\"facility\",\"02/12/15\",\"distribute_between_01/05/12\",\"could_introduce\",\"without_adequate_separation_which\",\"potential_for_cross_contamination\",\"compound\",\"fda_inspection_identify_gmp\",\"violation_potentially_impact\",\"initiate\",\"labeling_label_mixup\",\"concern_associate\",\"drug\",\"sterile\",\"within_expiry\",\"form\",\"find\",\"all_sterile_human\",\"compound_preservative_free_methylprednisolone\",\"skin_abscess_potentially_link\",\"firm_receive_seven_report\",\"adverse_reaction\",\"80mg/ml_10_ml_vial\",\"market_without_an_approved\",\"recent_fda_inspection\",\"potentially_mislabeled_as\",\"nda/anda\",\"follow_drug\",\"process\",\"specification\",\"ndc\",\"may_have_potentially\",\"quality_control_procedure_that\",\"concern_regard_quality_control\",\"fda_inspection_finding_result\",\"label\",\"use\",\"cgmp_deviation\",\"franck_'s_lab_inc.\",\"fungal_growth\",\"drugs_distribute_between_11/21/2011\",\"particulate_matter\",\"microorganism\",\"clean_room_where_sterile\",\"prepared\",\"that_present\",\"risk\",\"certain_quality_control_procedure\",\"contain\",\"pharmacy\",\"observe_during\",\"observation_associate\",\"compound_sterile_preparation\",\"compound_by\",\"capsule\",\"result\",\"stability_data_do_not\",\"subpotent_drug\",\"specification_result\",\"present\",\"mislabeled_as\",\"non_sterile\",\"conduct\",\"potential_risk\",\"voluntary\",\"firm\",\"microbial_contamination\",\"pharmacy_that\",\"bottle\",\"reveal\",\"connection\",\"martin_avenue_pharmacy_inc.\",\"impact\",\"hcl\",\"50\",\"fda_inspectional_finding_result\",\"can_not\",\"quality_control_process\",\"labeling:label_mixup\",\"mislabeled_as_one\",\"not_expire_due\",\"distribute_by_this\",\"environmental_sampling_reveal\",\"05/21/2012_because_fda\",\"non\",\"05/21/2012\",\"10\",\"fda_environmental_sampling\",\"syringe\",\"test\",\"assure\",\"potency\",\"labeling\",\"500\",\"vial\",\"manufacture\",\"glass_particulate\",\"products\",\"5\",\"testing\",\"fda\",\"also_cgmp_deviation\",\"25\",\"20\",\"100\",\"store\",\"injectable_drug\",\"customer_complaint\",\"date\",\"particulate_matter_api_contaminate\",\"produce_sterile\",\"fail_dissolution_specifications\",\"impurity\",\"manufacturer\",\"processing_controls\",\"identify_as\",\"potential\",\"drug_package\",\"complaint\",\"active_ingredient\",\"dissolution\",\"foreign_substance\",\"1\",\"inspection_observation_associate\",\"labeling_incorrect_or_missing\",\"support_expiry_recent\",\"solution\",\"fda_inspection\",\"support_expiry_potential_loss\",\"produce\",\"package\",\"contamination\",\"dietary_supplement\",\"affect\",\"mcg\",\"condition_potentially\",\"aseptic_practice\",\"their_compound\",\"observe\",\"fda_inspection_reveal_poor\",\"200_mg\",\"report\",\"ingredient\",\"2\",\"fail_impurities/degradation_specification\",\"fail\",\"potential_for\",\"not_assure\",\"chemical_contamination\",\"injection\",\"compound_human\",\"fda_approve\",\"same_environment\",\"veterinary\",\"same_practice_as_another\",\"exp_5/20/2014\",\"unexpired_sterile\",\"exp_5/1/2014\",\"do_not_adequately_investigate\",\"supplier_due\",\"active_ingredient_that\",\"specification_oos_result\",\"cgmp_deviation_firm\",\"stability\",\"make_use_an\",\"1/2\",\"subject\",\"particle\",\"specifications\",\"usp\",\"expiration_date\",\"label_mixup\",\"exp\",\"and/or_exp\",\"unit\",\"underdosed_or_have\",\"labeling_label_mix_up\",\"phenolphthalein\",\"regime\",\"an_incorrect_dosage\",\"find_concern\",\"unexpired\",\"fail_impurities/degradation_specifications_out\",\"leak\",\"package_insert\",\"exp_5/9/2014\",\"concern\",\"defective_container\",\"active_pharmaceutical_ingredient\",\"miss\",\"exp_5/22/2014\",\"assay\",\"exp_5/16/2014\",\"exp_5/15/2014\",\"contaminate\",\"labeling_label_error_on\",\"unapproved_new_drug\",\"superpotent\",\"poor_sterile\",\"defective_delivery_system\",\"production_practice_result\",\"for_their_finish\",\"exp_5/29/2014\",\"sibutramine\",\"potentially_contaminate\",\"30\",\"exp_5/30/2014\",\"superpotent_drug\",\"medication\",\"particulate\",\"injectable\",\"250\",\"for_assay\",\"40\",\"point\",\"levothyroxine_sodium_tablet\",\"relate\",\"guarantee\",\"quality_control_procedure\",\"an_unapproved_drug\",\"foreign\",\"glass\",\"carton\",\"during_stability_testing\",\"do_not_meet\",\"sildenafil\",\"exp_6/26/2014\",\"tablets\",\"release\",\"container\",\"fail_impurities/degradation_specifications\",\"distribute\",\"voluntarily\",\"number\",\"fill\",\"inc.\",\"failure\",\"'s\",\"time\",\"fail_tablet/capsule_specifications\",\"support_expiry\",\"declared_strength\",\"seal\",\"2.5_mg\",\"time_point\",\"fail_stability\",\"contain_undeclared_sibutramine\",\"identify\",\"api\",\"capsule_1000_mg\",\"er\",\"certain\",\"market_as\",\"documentation\",\"bag\",\"instead\",\"fail_dissolution\",\"foreign_tablets/capsule\",\"0.5\",\"receive\",\"exp_5/21/2014\",\"incorrect\",\"proper_environmental_monitoring_during\",\"150\",\"subpotent\",\"undeclared\",\"firm_'s_contract\",\"crystallization\",\"omega-3_fatty_acid\",\"contain_undeclared\",\"embed\",\"hcl_capsule\",\"potentially\",\"exp_5/28/2014\",\"unapproved_drug\",\"12_month\",\"sodium\",\"sterile_ophthalmic\",\"raw_material\",\"60\",\"tube\",\"12.5\",\"have_compromise\",\"sterility:all_sterile\",\"rohto\",\"may_have\",\"0\",\"process_deficiency\",\"80\",\"docusate_sodium_capsule_250\",\"that_could\",\"strength\",\"glass_vial\",\"laboratory\",\"fail_content_uniformity\",\"capsule_100_mg\",\"small\",\"limit\",\"ml\",\"correct\",\"stability_testing\",\"acid\",\"exp_6/12/2014\",\"325\",\"obtain\",\"fail_tablet/capsule_specification\",\"testing_lab\",\"an_incorrect\",\"15\",\"level\",\"acetaminophen\",\"cyanocobalamin\",\"indicate\",\"withdraw_from\",\"exp_5/17/2014\",\"may_not_meet\",\"fail_impurity/degradation_specification\",\"an_unapproved_new_drug\",\"guaifenesin\",\"exp_5/31/2014\",\"4\",\"aspirin\",\"prescription\",\"exp_6/27/2014\",\"reliable\",\"guaifenesin_er_tablet_600\",\"exp_5/13/2014\",\"concerned_test_result_obtain\",\"taint\",\"low\",\"cholecalciferol_tablet\",\"select_sterile\",\"laboratory_result\",\"make_this\",\"impurities/degradation\",\"not_affect\",\"chew_tablet\",\"exp_6/6/2014\",\"exp_6/25/2014\",\"stability_datum_do_not\",\"precipitate\",\"propranolol_hcl\",\"multivitamin/multimineral\",\"break\",\"stability_failure\",\"preservative\",\"stainless_steel\",\"do_not\",\"good_manufacturing_practices\",\"calcium\",\"discoloration\",\"approve_nda/anda\",\"substance\",\"insufficient_datum\",\"dr\",\"salicylic_acid\",\"stability_time_point\",\"subpotent_single_ingredient\",\"lozenge\",\"previous_lot_there\",\"determine_that_other\",\"burkholderia_cepacia\",\"market_without\",\"quality_review\",\"tadalafil\",\"manufacturing_firm\"],\"word\":[\"sterility\",\"assurance\",\"product\",\"lack\",\"penicillin\",\"recall\",\"tablet\",\"repackaged\",\"lot\",\"pedigree\",\"presence\",\"mg_ndc\",\"mg\",\"cross_contamination\",\"recall_because_they\",\"quality\",\"facility\",\"02/12/15\",\"distribute_between_01/05/12\",\"could_introduce\",\"without_adequate_separation_which\",\"potential_for_cross_contamination\",\"compound\",\"fda_inspection_identify_gmp\",\"violation_potentially_impact\",\"initiate\",\"labeling_label_mixup\",\"concern_associate\",\"drug\",\"sterile\",\"within_expiry\",\"form\",\"find\",\"all_sterile_human\",\"compound_preservative_free_methylprednisolone\",\"skin_abscess_potentially_link\",\"firm_receive_seven_report\",\"adverse_reaction\",\"80mg/ml_10_ml_vial\",\"market_without_an_approved\",\"recent_fda_inspection\",\"potentially_mislabeled_as\",\"nda/anda\",\"follow_drug\",\"process\",\"specification\",\"ndc\",\"may_have_potentially\",\"quality_control_procedure_that\",\"concern_regard_quality_control\",\"fda_inspection_finding_result\",\"label\",\"use\",\"cgmp_deviation\",\"franck_'s_lab_inc.\",\"fungal_growth\",\"drugs_distribute_between_11/21/2011\",\"particulate_matter\",\"microorganism\",\"clean_room_where_sterile\",\"prepared\",\"that_present\",\"risk\",\"certain_quality_control_procedure\",\"contain\",\"pharmacy\",\"observe_during\",\"observation_associate\",\"compound_sterile_preparation\",\"compound_by\",\"capsule\",\"result\",\"stability_data_do_not\",\"subpotent_drug\",\"specification_result\",\"present\",\"mislabeled_as\",\"non_sterile\",\"conduct\",\"potential_risk\",\"voluntary\",\"firm\",\"microbial_contamination\",\"pharmacy_that\",\"bottle\",\"reveal\",\"connection\",\"martin_avenue_pharmacy_inc.\",\"impact\",\"hcl\",\"50\",\"fda_inspectional_finding_result\",\"can_not\",\"quality_control_process\",\"labeling:label_mixup\",\"mislabeled_as_one\",\"not_expire_due\",\"distribute_by_this\",\"environmental_sampling_reveal\",\"05/21/2012_because_fda\",\"non\",\"05/21/2012\",\"10\",\"fda_environmental_sampling\",\"syringe\",\"test\",\"assure\",\"potency\",\"labeling\",\"500\",\"vial\",\"manufacture\",\"glass_particulate\",\"products\",\"5\",\"testing\",\"fda\",\"also_cgmp_deviation\",\"25\",\"20\",\"100\",\"store\",\"injectable_drug\",\"customer_complaint\",\"date\",\"particulate_matter_api_contaminate\",\"produce_sterile\",\"fail_dissolution_specifications\",\"impurity\",\"manufacturer\",\"processing_controls\",\"identify_as\",\"potential\",\"drug_package\",\"complaint\",\"active_ingredient\",\"dissolution\",\"foreign_substance\",\"1\",\"inspection_observation_associate\",\"labeling_incorrect_or_missing\",\"support_expiry_recent\",\"solution\",\"fda_inspection\",\"support_expiry_potential_loss\",\"produce\",\"package\",\"contamination\",\"dietary_supplement\",\"affect\",\"mcg\",\"condition_potentially\",\"aseptic_practice\",\"their_compound\",\"observe\",\"fda_inspection_reveal_poor\",\"200_mg\",\"report\",\"ingredient\",\"2\",\"fail_impurities/degradation_specification\",\"fail\",\"potential_for\",\"not_assure\",\"chemical_contamination\",\"injection\",\"compound_human\",\"fda_approve\",\"same_environment\",\"veterinary\",\"same_practice_as_another\",\"exp_5/20/2014\",\"unexpired_sterile\",\"exp_5/1/2014\",\"do_not_adequately_investigate\",\"supplier_due\",\"active_ingredient_that\",\"specification_oos_result\",\"cgmp_deviation_firm\",\"stability\",\"make_use_an\",\"1/2\",\"subject\",\"particle\",\"specifications\",\"usp\",\"expiration_date\",\"label_mixup\",\"exp\",\"and/or_exp\",\"unit\",\"underdosed_or_have\",\"labeling_label_mix_up\",\"phenolphthalein\",\"regime\",\"an_incorrect_dosage\",\"find_concern\",\"unexpired\",\"fail_impurities/degradation_specifications_out\",\"leak\",\"package_insert\",\"exp_5/9/2014\",\"concern\",\"defective_container\",\"active_pharmaceutical_ingredient\",\"miss\",\"exp_5/22/2014\",\"assay\",\"exp_5/16/2014\",\"exp_5/15/2014\",\"contaminate\",\"labeling_label_error_on\",\"unapproved_new_drug\",\"superpotent\",\"poor_sterile\",\"defective_delivery_system\",\"production_practice_result\",\"for_their_finish\",\"exp_5/29/2014\",\"sibutramine\",\"potentially_contaminate\",\"30\",\"exp_5/30/2014\",\"superpotent_drug\",\"medication\",\"particulate\",\"injectable\",\"250\",\"for_assay\",\"40\",\"point\",\"levothyroxine_sodium_tablet\",\"relate\",\"guarantee\",\"quality_control_procedure\",\"an_unapproved_drug\",\"foreign\",\"glass\",\"carton\",\"during_stability_testing\",\"do_not_meet\",\"sildenafil\",\"exp_6/26/2014\",\"tablets\",\"release\",\"container\",\"fail_impurities/degradation_specifications\",\"distribute\",\"voluntarily\",\"number\",\"fill\",\"inc.\",\"failure\",\"'s\",\"time\",\"fail_tablet/capsule_specifications\",\"support_expiry\",\"declared_strength\",\"seal\",\"2.5_mg\",\"time_point\",\"fail_stability\",\"contain_undeclared_sibutramine\",\"identify\",\"api\",\"capsule_1000_mg\",\"er\",\"certain\",\"market_as\",\"documentation\",\"bag\",\"instead\",\"fail_dissolution\",\"foreign_tablets/capsule\",\"0.5\",\"receive\",\"exp_5/21/2014\",\"incorrect\",\"proper_environmental_monitoring_during\",\"150\",\"subpotent\",\"undeclared\",\"firm_'s_contract\",\"crystallization\",\"omega-3_fatty_acid\",\"contain_undeclared\",\"embed\",\"hcl_capsule\",\"potentially\",\"exp_5/28/2014\",\"unapproved_drug\",\"12_month\",\"sodium\",\"sterile_ophthalmic\",\"raw_material\",\"60\",\"tube\",\"12.5\",\"have_compromise\",\"sterility:all_sterile\",\"rohto\",\"may_have\",\"0\",\"process_deficiency\",\"80\",\"docusate_sodium_capsule_250\",\"that_could\",\"strength\",\"glass_vial\",\"laboratory\",\"fail_content_uniformity\",\"capsule_100_mg\",\"small\",\"limit\",\"ml\",\"correct\",\"stability_testing\",\"acid\",\"exp_6/12/2014\",\"325\",\"obtain\",\"fail_tablet/capsule_specification\",\"testing_lab\",\"an_incorrect\",\"15\",\"level\",\"acetaminophen\",\"cyanocobalamin\",\"indicate\",\"withdraw_from\",\"exp_5/17/2014\",\"may_not_meet\",\"fail_impurity/degradation_specification\",\"an_unapproved_new_drug\",\"guaifenesin\",\"exp_5/31/2014\",\"4\",\"aspirin\",\"prescription\",\"exp_6/27/2014\",\"reliable\",\"guaifenesin_er_tablet_600\",\"exp_5/13/2014\",\"concerned_test_result_obtain\",\"taint\",\"low\",\"cholecalciferol_tablet\",\"select_sterile\",\"laboratory_result\",\"make_this\",\"impurities/degradation\",\"not_affect\",\"chew_tablet\",\"exp_6/6/2014\",\"exp_6/25/2014\",\"stability_datum_do_not\",\"precipitate\",\"propranolol_hcl\",\"multivitamin/multimineral\",\"break\",\"stability_failure\",\"preservative\",\"stainless_steel\",\"do_not\",\"good_manufacturing_practices\",\"calcium\",\"discoloration\",\"approve_nda/anda\",\"substance\",\"insufficient_datum\",\"dr\",\"salicylic_acid\",\"stability_time_point\",\"subpotent_single_ingredient\",\"lozenge\",\"previous_lot_there\",\"determine_that_other\",\"burkholderia_cepacia\",\"market_without\",\"quality_review\",\"tadalafil\",\"manufacturing_firm\"],\"x_coord\":[-8.427880789001843,7.898232701125545,-20.207018767464806,-26.316070482248605,47.524377943840086,-16.230945845260212,10.495605936454819,52.693222794051856,-3.4622132358000233,16.59155444916776,-5.698254612322265,25.422888421671075,13.115153621638568,34.508002291344084,49.5817412468766,23.93735616938305,53.07661506866452,50.248734380258234,47.465144977618436,47.929179912522756,53.08864148744211,49.91787969971838,19.919028735015143,22.03270711608873,25.892538726590484,-8.226395175569062,36.62902725203923,33.63119354583505,-17.125294291834496,37.498310136560455,37.10307837707518,20.194413073485222,-37.42007643422356,11.523501620714796,10.701093992143154,17.039925286510112,22.725841139854925,18.77900047217412,14.558472194211367,-16.958366972411966,31.39436184997123,12.362642500282757,-19.127584751013714,25.128859399871843,-44.03374635168472,-47.455786226820784,7.888646066508409,17.945438630059623,37.52988433088752,-48.39170531521668,-61.42228079816551,-9.311747215034856,-19.307829182866378,-12.565100290273486,35.482053879194886,16.021275454763806,38.44922024026168,-1.2966677428361875,15.76395204271931,37.093411042633214,37.86862059397059,17.771272776120412,17.336806964566513,15.042911881402176,38.11770241058829,22.691869156154983,28.645557524031698,12.297528121829506,25.166843953503356,44.247942852599124,-2.875425153503274,-25.265192369131068,20.680616265821428,-47.0695776268208,-54.38184463633619,4.319500051176563,53.30205619212993,57.62145253606414,-77.1475983113979,67.34872904395507,29.56454443502116,-17.530484804712387,50.41791499493345,59.343063139904906,21.06404584209036,19.834789909935864,17.82911075688672,-15.20007061067599,33.9362501181983,42.26787316405844,5.965345117149951,36.64521397307913,13.967327426560052,18.516260919515844,-25.115146440092307,23.955363530156784,41.034627016743315,29.790390328840164,28.136284686354372,31.83057298356939,0.5951450840405932,34.34840828199486,22.856176002453264,41.2972786159376,-6.273307684269975,-34.56770657753287,5.633583869794574,-2.3022890029807357,-2.2239922356209125,13.730340786432711,-30.856251809151562,-14.396365379266534,-4.210772757291938,24.424507646421038,49.563427047013334,-43.82282839536716,-18.07126281991175,25.17563087974672,-6.825597275154448,38.241723676869555,4.816386842776734,-3.8538861304271013,-5.992309007502634,-23.016473237878298,8.04934316470306,-6.5518051094817755,-4.486620069890327,-77.33226862997462,-33.168862475160566,-9.559375516637246,4.489118546476779,-25.10604934307487,-1.6859178401448562,-1.952257675156932,-47.432776132491284,26.165108777236856,-11.98005529229008,18.745325599385954,-40.19293546498842,21.762892952623098,22.832594519740287,-32.989290284504754,4.165750004805594,3.201788521010455,-36.8304566599907,-3.692824198226839,-26.6345906843196,-17.380561467247094,-28.003407054975675,37.74981882027083,30.020642603277487,-7.222538693368082,38.40871582680743,35.639774849819815,35.52179798023711,10.208903713907084,3.5484267669914957,-28.698005232335895,24.25396151416334,-11.163658303224665,-43.086319845501386,-30.01252725856443,-50.28714993525944,25.985875960385723,-19.845376491565066,-25.311052330107813,57.178717632400165,-13.35505076350427,45.1495442466895,60.95007508582686,50.56833907059893,52.51432959758844,19.073990829159996,31.774340553021407,-51.54709586784271,37.96437844030148,26.74134194233495,-51.144666207802736,60.910693711283336,45.66687186004086,7.423156673143476,30.426434148431255,35.42455764133079,-30.718592149041246,5.960324244414198,9.088002661755729,29.267333942419302,27.427539894825163,6.923574020457869,-3.8121568258792813,9.876573096414825,-3.2700864863408237,52.04140691346654,-23.72313716858875,51.527195668418095,55.41704730757857,-27.47947316053039,1.5309616615597699,-43.7553882319466,-16.522931184674977,23.12098229216051,-18.118848604706493,-31.219386314276445,2.736261617157247,6.012373225214441,-12.16388518478907,-27.984627698017785,16.098609197540174,21.35848232954594,37.865005111544434,-1.9898408288438438,22.444865165729258,11.368791502270597,-42.89542353257019,36.87702126791488,-20.13619353331924,17.196584452115047,-21.611443919112,-21.69466632880287,41.0962456821108,42.66791948231306,34.83434561559611,-38.662438421936635,-21.7865966880981,27.93049967594762,4.509221222536949,-44.442099493470124,-41.43706482485372,29.860633634503664,7.622833126666343,-60.65997521441866,23.504082238126227,-1.68285197600561,12.905497306114393,-35.66903396642016,-10.9388789177008,-15.83061804706973,-11.74840156570092,-11.432962224935105,-37.62808171228198,-8.396476899010715,-42.562217827155855,33.821299256026585,-33.72394897973687,-35.13744858826308,-42.560089382418255,-1.805815413316549,-14.112760089552992,11.625016551409516,-27.47119711284981,-43.684545609458745,-30.722690381328327,-5.261090101698644,-27.36896025433599,-17.654110722128763,-2.3495418197042013,-27.148304855232283,-18.66380208294902,-45.76522923661217,30.73980789108628,-38.199612256396605,-17.497025363168078,-71.29221863620374,1.074504746730591,-0.7491993370605833,11.41701952908749,-48.038619385042004,-29.342711148864872,-35.955126490307244,-8.924406416458996,-7.013132014208125,34.17184969833918,-11.671081558069828,17.240232109256258,-38.24705672628571,-10.127386137624457,33.982706484735694,-45.90915875879334,-17.699462000199418,-54.72220454166507,39.711066720542355,-16.63351027671126,42.04223540892584,-56.12203921123098,-16.852115409522035,-20.3416120627547,-20.844893270479847,20.23586706601711,17.125307774917065,18.18154604893926,-24.96523119407712,7.4529774363372825,-41.66594091964853,15.183112746810476,-14.436949224621156,15.933197129011422,14.094937980953635,29.881846542817858,-7.407592783819334,49.58698016521175,37.592642637580184,-7.501649852768551,6.913965890931759,8.793367270732888,27.884793622053042,20.115715386248045,-40.27977692000294,-24.208053903900584,22.832963117720016,-6.599538429696668,-51.78672962474414,3.736295833030691,2.7472399283175233,-46.19536963240873,-41.179669339658275,-5.762374500694633,-14.096640361507452,-36.702231280068084,0.26076808301174653,-26.232636994202316,11.319743942570199,-22.672473222536947,-39.38900199410769,-7.232362240760556,12.66601921456143,-16.241735491871587,-3.9096798449031684,41.53426783835984,9.684583769041152,16.611187727994896,-19.787166035920006,23.44293369799894,25.27694483981401,-15.271530095442166,-11.689589523204152,34.888999867291055,46.27433487951771,-40.48066281702772,-9.325825880518993,11.765319985381963,24.47556380160051,-22.26704089719214,-35.12256215644061,-11.14548406835478,17.19587161989324,34.020993347162175,-14.425284758353905,13.068341442713113,9.500815917257182,-47.97583906244042,27.63216334924993,-16.968487187441024,-2.0503769815918558,34.964602427482426,-24.892177126555577,-27.788983556555827,-47.047486567381995,-38.0339764956833,26.607817422136137,-37.86329655713738,-9.285731548232548,-48.40177652352432,-26.872010755740146,-45.78164442397661,-16.463516538479706,-3.1124449700087182,-23.20598913141027,2.8920907341274926,-46.08550673343035,22.89207325211825,-36.040130228363495,19.340379109830483,-44.58212126212013,-16.160516815852557,32.510512330311926,-29.94850993349796,0.8430744234221287,-22.755448680051792,-44.1430986979487,81.97212496162301,-21.185771272017146,-25.338497317646414],\"y_coord\":[15.936145426625341,-1.578057242305833,2.5627141097463166,3.6740489670784737,11.912825746930263,12.898171759213282,-20.07425278473817,8.2406719612047,16.510038269808742,-40.06182907122913,19.552289830038355,-37.8995308245251,-40.39131347378055,-9.52419120755314,6.020968492505546,37.08152889343328,10.26059472228524,17.96793325783106,17.48390652439095,9.587073528152317,15.294817490389251,13.423234292225455,34.762726531876325,37.8366396094919,35.67269061830634,-0.3828152664420492,-47.92434793616722,8.82434154253377,-6.521044488454566,16.066610753742268,-2.839718009936321,52.68634045977722,75.09671652516404,52.235998601437885,61.94350748792209,57.01663512676048,51.40410905377912,56.58329464477298,49.02418369009888,-35.32319495595992,-16.992114465706305,-54.996672081162544,-34.02405603366546,-54.20617318726318,33.291457912001356,4.358323651157249,-40.89849698841061,-62.111320606420335,6.850604226334019,29.160355638753817,32.02805056256887,45.74588048609173,2.211381635243511,-7.7687189318646,26.985963979138045,39.852363383913996,26.15771192967615,36.25782541744687,37.503212877629416,24.211055757458567,23.047963634783695,-3.3035051131204805,-7.669884890224416,-5.504859874403148,-6.558166719713277,43.93271070708649,8.691524127638093,18.853522948577204,24.183383538382916,21.816340530598286,-32.24084705767015,11.199902185721129,-0.32669942634746985,-17.852374093572877,-3.244931946499574,-0.44253404196324997,-44.23547821452381,14.121110416409842,25.334617041326474,2.503255336576776,56.10759980034261,35.47165569865563,-22.718061144590564,53.46642814587412,11.169208081902445,21.446704273788846,4.875548904315725,33.00769333030545,2.906587713921614,-9.835761009717077,-27.310751768748293,10.894623365876178,27.850675728073764,2.0461444891237495,13.75221594081634,-59.406090641203434,31.210589671085962,29.86799435299773,16.0445042792593,23.921849792722952,12.657795466667293,29.426566911864416,29.092237040451426,-51.80016363697649,-27.042634071683047,-1.268210865920949,5.007050575035672,-11.450269609092144,49.43693185346352,-29.062023623019368,40.78996220841544,4.294639883547315,-48.54881570652022,-7.337997619891865,-27.77125353608171,-9.33226359929192,-23.18810388741071,-19.43550122496995,12.069728791891167,-26.709102484759775,-22.396561305076226,-28.248518043390426,-53.462249602997666,19.431694708642382,47.826789569215954,-44.653890784434694,-44.30492103126429,30.98308128813435,5.8801790817213035,21.728278961962367,15.909509930453444,35.359506253669565,-6.808014759222549,-16.81749008879979,21.673893017145694,-14.169421667894435,-28.466018662935028,15.154892593727133,20.534948758217407,-0.08998797858942123,0.7069513753574832,33.54112172536341,39.97420067933419,-10.06015223452719,33.626389863131415,-19.194401765074716,21.55399089930787,55.357129734324495,-39.75760838515625,39.9916018258907,51.331470867217035,8.362331496653871,45.33502325104889,34.00768617443208,42.49296992738046,-40.21620498087336,-40.19238777250548,20.42545087588191,-27.787631937397116,-18.386894483632133,9.116543150025674,44.16646674614963,-3.597900171949802,40.14888952290939,50.254730808107965,26.092465486321174,22.92829015813757,-31.387401541771652,29.431482085665493,24.783222539594366,24.406624011718023,28.027467930172485,-46.11037810755504,-53.38910073911235,0.14830631197327016,-9.352700547450354,0.6694473391388396,-6.863304590405158,-36.02840644135855,-33.8250183095139,-49.607587166861755,-5.289769371179658,19.387365837049582,-7.478961422165979,27.965660839330273,-25.49374718096833,-57.686423870069675,-39.98486595765292,45.17443049120236,32.964387287292816,49.90876460032298,41.807999779649556,-18.058852479883207,-26.244717895490204,-14.60392190110572,-14.85712936693731,-6.2226118864821105,1.136215455930751,-25.65749868145003,-2.4098783910878683,-32.89146280042735,-14.74522978944206,27.68069079064993,53.60379357093646,39.99100783185319,47.39958631337892,6.157508666817931,-45.71525962631669,-54.00129741552783,-14.422204340568932,59.35728369758874,-44.38866819662692,10.706231600899898,-32.50508116182272,-20.068773044900528,10.631336468776801,17.865090467353927,-29.00715581468406,-24.028554755548257,-2.4650530485895095,-21.799621198690883,-56.62833950725494,34.756118757831025,33.389414966520754,-49.353503211033306,10.275125303557376,2.402170690151993,-11.68773665718388,-23.91828943290568,-37.44114298154387,-6.750768845188517,-43.379332164379484,17.748078328421762,1.3781768523049385,-20.629420117965775,40.106524571391546,57.253798732022645,30.00602817387329,-15.2572651123149,0.020592219467144957,-37.816189793430425,-5.9965434906086825,-40.42832783107133,16.1905597611223,23.291584638051436,8.532116669016578,25.62848545951094,43.3840149606498,-12.087807349674266,13.995546848400103,39.23411957066977,6.990242738210365,43.48860966961282,30.238048761186548,16.390394760430258,23.781268985432924,-26.168177763360152,-10.525704371613593,-4.8563652957171035,-43.319779949094595,-8.702932268892805,41.42875187227645,-52.3484749009935,-2.752896459407075,58.41652798798951,-42.79766353025423,-9.951536295124347,-31.856657403284927,44.57870282007952,49.62891629303525,23.174498468225373,-36.40014719979876,37.23323817809846,-53.318248103417744,-15.837745787089231,52.69165135905432,-17.877154002067893,21.458898105151615,19.610728215159877,33.4543640680708,-35.37699463504374,-30.221788567190693,-15.454935868764252,12.47930826545831,-57.240895758156995,-36.169034477363795,27.350555291896978,-48.79213036104758,-21.380199983463438,-17.898199792867924,-35.358267412845514,-47.95440616736127,29.344211712131717,31.50586740994251,20.143429756254946,-22.27141522693533,-15.3444258987934,-30.417243684877313,33.39176372569025,34.57105532804083,-22.88906907082475,-8.154467476255846,23.234713089253216,22.010085981608608,-41.22372714689168,27.37387873519137,-4.8019532456288925,7.364484858177158,-41.52871350907103,50.03842725501915,-12.511836066188751,-3.4675768273928336,-31.698548532789278,0.49323739105596476,-1.0658801487482605,52.565146971766225,50.3431093921301,43.7343321700327,-27.369801681855428,-12.559871261753049,-53.81102806160171,-24.14825460034042,-8.034810834676636,47.354805860134064,-31.644066973590505,-51.28530654038909,31.780977251516394,-38.594703738526086,-7.863088232436483,-68.62439307987623,-18.78618113266742,-32.87315409280489,-45.351801013641,13.492272682221868,-10.805011740621092,-6.966893999622978,-3.3148360143050044,2.8721342166532065,-16.178477420327603,-53.44406300935138,15.027403705740655,-18.09351100972599,10.772981018452736,1.7924084626458776,-28.59141862470931,-47.43190964506567,-56.00038276366197,-34.70723568437253,-47.29073217894402,-31.618289841838504,-30.133336941548322,-59.02891581008849,10.705475200502823,-43.87142448274506,-21.366459399375067,0.6603532745965437,-36.43038583676713,8.738931878559372,-27.064275495161194,-32.227685442721665,43.50902405522636,-39.327867731323366,-28.41383635533112,-33.63606441051337,-28.239952876639585,-2.683688940307569,-31.98885748157299,-45.592627142877866,-7.518858034388986,8.409236995596096,25.5712655763333,-27.38378105477478,-37.913308616429205,46.766934529249134,-36.35638119482711,15.7518842554396,-43.40423760057459,22.933274704793575,53.3973247032409,-8.68883699939283,-6.728642726970583,-38.17495329745346]}},\"id\":\"8ab3186b-9483-460b-b380-a48a6b2292ae\",\"type\":\"ColumnDataSource\"},{\"attributes\":{\"callback\":null},\"id\":\"0a89d6b5-9c7a-4695-881e-db58ae17bde9\",\"type\":\"DataRange1d\"},{\"attributes\":{\"active_drag\":\"auto\",\"active_scroll\":{\"id\":\"ec665a16-cfec-4cbe-a6ef-6d2260a020a4\",\"type\":\"WheelZoomTool\"},\"active_tap\":\"auto\",\"tools\":[{\"id\":\"d306a1a8-b909-4fd2-8b21-26a72e0d6eea\",\"type\":\"PanTool\"},{\"id\":\"ec665a16-cfec-4cbe-a6ef-6d2260a020a4\",\"type\":\"WheelZoomTool\"},{\"id\":\"c8053077-04e9-4328-954c-e3309cad6011\",\"type\":\"BoxZoomTool\"},{\"id\":\"051447f1-c457-4aa6-8316-e0f83b0eff6d\",\"type\":\"BoxSelectTool\"},{\"id\":\"d3979c2a-9557-4615-a687-3682c33e2119\",\"type\":\"ResizeTool\"},{\"id\":\"ca48d166-0e93-47e1-a376-37f32dafd4a0\",\"type\":\"ResetTool\"},{\"id\":\"73a8abce-1f48-42e5-9800-f6e5b7730e28\",\"type\":\"HoverTool\"}]},\"id\":\"f10872fd-a808-43c7-bf37-2c5bd9589f0a\",\"type\":\"Toolbar\"},{\"attributes\":{\"plot\":{\"id\":\"7c81ad3e-90a5-4aae-ac9f-81c66a17c864\",\"subtype\":\"Figure\",\"type\":\"Plot\"}},\"id\":\"ca48d166-0e93-47e1-a376-37f32dafd4a0\",\"type\":\"ResetTool\"},{\"attributes\":{\"formatter\":{\"id\":\"3a092e41-acd8-4816-b97f-0e071b84fff9\",\"type\":\"BasicTickFormatter\"},\"plot\":{\"id\":\"7c81ad3e-90a5-4aae-ac9f-81c66a17c864\",\"subtype\":\"Figure\",\"type\":\"Plot\"},\"ticker\":{\"id\":\"1a7a5043-8345-44d8-bfeb-01357fd77695\",\"type\":\"BasicTicker\"},\"visible\":false},\"id\":\"fb5c7de4-3ca4-4cfd-857b-efbf045de68f\",\"type\":\"LinearAxis\"},{\"attributes\":{\"dimension\":1,\"grid_line_color\":{\"value\":null},\"plot\":{\"id\":\"7c81ad3e-90a5-4aae-ac9f-81c66a17c864\",\"subtype\":\"Figure\",\"type\":\"Plot\"},\"ticker\":{\"id\":\"1a7a5043-8345-44d8-bfeb-01357fd77695\",\"type\":\"BasicTicker\"}},\"id\":\"dc564fe1-78fa-42fb-ba0d-bee1bf0d3225\",\"type\":\"Grid\"},{\"attributes\":{\"plot\":{\"id\":\"7c81ad3e-90a5-4aae-ac9f-81c66a17c864\",\"subtype\":\"Figure\",\"type\":\"Plot\"}},\"id\":\"d3979c2a-9557-4615-a687-3682c33e2119\",\"type\":\"ResizeTool\"},{\"attributes\":{\"data_source\":{\"id\":\"8ab3186b-9483-460b-b380-a48a6b2292ae\",\"type\":\"ColumnDataSource\"},\"glyph\":{\"id\":\"868c0f87-d4e1-4019-9976-0902f4f26d35\",\"type\":\"Circle\"},\"hover_glyph\":{\"id\":\"93797eac-3d20-467b-8474-f5c85a86cd6d\",\"type\":\"Circle\"},\"nonselection_glyph\":{\"id\":\"9ef5454f-f7b8-453e-9f0f-1a95c4510330\",\"type\":\"Circle\"},\"selection_glyph\":null},\"id\":\"c5b7dc8a-59ff-4f57-a1e7-4be678d01e5a\",\"type\":\"GlyphRenderer\"},{\"attributes\":{\"plot\":{\"id\":\"7c81ad3e-90a5-4aae-ac9f-81c66a17c864\",\"subtype\":\"Figure\",\"type\":\"Plot\"}},\"id\":\"ec665a16-cfec-4cbe-a6ef-6d2260a020a4\",\"type\":\"WheelZoomTool\"},{\"attributes\":{},\"id\":\"2903f9f7-248d-4fd6-aa90-e166da048a2c\",\"type\":\"BasicTickFormatter\"},{\"attributes\":{\"plot\":{\"id\":\"7c81ad3e-90a5-4aae-ac9f-81c66a17c864\",\"subtype\":\"Figure\",\"type\":\"Plot\"}},\"id\":\"d306a1a8-b909-4fd2-8b21-26a72e0d6eea\",\"type\":\"PanTool\"},{\"attributes\":{},\"id\":\"3a092e41-acd8-4816-b97f-0e071b84fff9\",\"type\":\"BasicTickFormatter\"},{\"attributes\":{\"overlay\":{\"id\":\"cdf2cfc2-3a5c-4644-bfc0-d0adc36bb556\",\"type\":\"BoxAnnotation\"},\"plot\":{\"id\":\"7c81ad3e-90a5-4aae-ac9f-81c66a17c864\",\"subtype\":\"Figure\",\"type\":\"Plot\"}},\"id\":\"c8053077-04e9-4328-954c-e3309cad6011\",\"type\":\"BoxZoomTool\"},{\"attributes\":{},\"id\":\"d95a48af-221a-4b1e-8983-71cd170770ce\",\"type\":\"BasicTicker\"},{\"attributes\":{\"fill_alpha\":{\"value\":0.1},\"fill_color\":{\"value\":\"blue\"},\"line_alpha\":{\"value\":0.2},\"line_color\":{\"value\":\"blue\"},\"size\":{\"units\":\"screen\",\"value\":10},\"x\":{\"field\":\"x_coord\"},\"y\":{\"field\":\"y_coord\"}},\"id\":\"868c0f87-d4e1-4019-9976-0902f4f26d35\",\"type\":\"Circle\"},{\"attributes\":{\"plot\":null,\"text\":\"t-SNE Word Embeddings\",\"text_font_size\":{\"value\":\"16pt\"}},\"id\":\"37945968-dfc6-46b7-94a6-2dfdf351bf18\",\"type\":\"Title\"},{\"attributes\":{},\"id\":\"1a7a5043-8345-44d8-bfeb-01357fd77695\",\"type\":\"BasicTicker\"}],\"root_ids\":[\"7c81ad3e-90a5-4aae-ac9f-81c66a17c864\"]},\"title\":\"Bokeh Application\",\"version\":\"0.12.4\"}};\n", | |
| " var render_items = [{\"docid\":\"0da6728f-c9ad-45fd-ad12-9bb446d72c06\",\"elementid\":\"1ffba516-d86c-45bc-9d9c-f02e7c30c12b\",\"modelid\":\"7c81ad3e-90a5-4aae-ac9f-81c66a17c864\"}];\n", | |
| " \n", | |
| " Bokeh.embed.embed_items(docs_json, render_items);\n", | |
| " };\n", | |
| " if (document.readyState != \"loading\") fn();\n", | |
| " else document.addEventListener(\"DOMContentLoaded\", fn);\n", | |
| " })();\n", | |
| " },\n", | |
| " function(Bokeh) {\n", | |
| " }\n", | |
| " ];\n", | |
| " \n", | |
| " function run_inline_js() {\n", | |
| " \n", | |
| " if ((window.Bokeh !== undefined) || (force === true)) {\n", | |
| " for (var i = 0; i < inline_js.length; i++) {\n", | |
| " inline_js[i](window.Bokeh);\n", | |
| " }if (force === true) {\n", | |
| " display_loaded();\n", | |
| " }} else if (Date.now() < window._bokeh_timeout) {\n", | |
| " setTimeout(run_inline_js, 100);\n", | |
| " } else if (!window._bokeh_failed_load) {\n", | |
| " console.log(\"Bokeh: BokehJS failed to load within specified timeout.\");\n", | |
| " window._bokeh_failed_load = true;\n", | |
| " } else if (force !== true) {\n", | |
| " var cell = $(document.getElementById(\"1ffba516-d86c-45bc-9d9c-f02e7c30c12b\")).parents('.cell').data().cell;\n", | |
| " cell.output_area.append_execute_result(NB_LOAD_WARNING)\n", | |
| " }\n", | |
| " \n", | |
| " }\n", | |
| " \n", | |
| " if (window._bokeh_is_loading === 0) {\n", | |
| " console.log(\"Bokeh: BokehJS loaded, going straight to plotting\");\n", | |
| " run_inline_js();\n", | |
| " } else {\n", | |
| " load_libs(js_urls, function() {\n", | |
| " console.log(\"Bokeh: BokehJS plotting callback run at\", now());\n", | |
| " run_inline_js();\n", | |
| " });\n", | |
| " }\n", | |
| " }(this));\n", | |
| "</script>" | |
| ] | |
| }, | |
| "metadata": {}, | |
| "output_type": "display_data" | |
| } | |
| ], | |
| "source": [ | |
| "# add our DataFrame as a ColumnDataSource for Bokeh\n", | |
| "plot_data = ColumnDataSource(tsne_vectors)\n", | |
| "\n", | |
| "# create the plot and configure the\n", | |
| "# title, dimensions, and tools\n", | |
| "tsne_plot = figure(title='t-SNE Word Embeddings',\n", | |
| " plot_width = 800,\n", | |
| " plot_height = 800,\n", | |
| " tools= ('pan, wheel_zoom, box_zoom,'\n", | |
| " 'box_select, resize, reset'),\n", | |
| " active_scroll='wheel_zoom')\n", | |
| "\n", | |
| "# add a hover tool to display words on roll-over\n", | |
| "tsne_plot.add_tools( HoverTool(tooltips = '@word') )\n", | |
| "\n", | |
| "# draw the words as circles on the plot\n", | |
| "tsne_plot.circle('x_coord', 'y_coord', source=plot_data,\n", | |
| " color='blue', line_alpha=0.2, fill_alpha=0.1,\n", | |
| " size=10, hover_line_color='black')\n", | |
| "\n", | |
| "# configure visual elements of the plot\n", | |
| "tsne_plot.title.text_font_size = value('16pt')\n", | |
| "tsne_plot.xaxis.visible = False\n", | |
| "tsne_plot.yaxis.visible = False\n", | |
| "tsne_plot.grid.grid_line_color = None\n", | |
| "tsne_plot.outline_line_color = None\n", | |
| "\n", | |
| "# engage!\n", | |
| "show(tsne_plot);" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "## Conclusion" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Whew! Let's round up the major components that we've seen:\n", | |
| "1. Text processing with **spaCy**\n", | |
| "1. Automated **phrase modeling**\n", | |
| "1. Topic modeling with **LDA** $\\ \\longrightarrow\\ $ visualization with **pyLDAvis**\n", | |
| "1. Word vector modeling with **word2vec** $\\ \\longrightarrow\\ $ visualization with **t-SNE**\n", | |
| "\n", | |
| "#### Why use these models?\n", | |
| "Dense vector representations for text like LDA and word2vec can greatly improve performance for a number of common, text-heavy problems like:\n", | |
| "- Text classification\n", | |
| "- Search\n", | |
| "- Recommendations\n", | |
| "- Question answering" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": null, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [] | |
| } | |
| ], | |
| "metadata": { | |
| "kernelspec": { | |
| "display_name": "Python 3", | |
| "language": "python", | |
| "name": "python3" | |
| }, | |
| "language_info": { | |
| "codemirror_mode": { | |
| "name": "ipython", | |
| "version": 3 | |
| }, | |
| "file_extension": ".py", | |
| "mimetype": "text/x-python", | |
| "name": "python", | |
| "nbconvert_exporter": "python", | |
| "pygments_lexer": "ipython3", | |
| "version": "3.5.2" | |
| } | |
| }, | |
| "nbformat": 4, | |
| "nbformat_minor": 0 | |
| } |
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment