Last active
December 7, 2019 18:08
-
-
Save DMTSource/521a84269087d9e21ad516193c0e566f to your computer and use it in GitHub Desktop.
Example on how to extract and plot fitness diversity for a population, hall of fame, or frontier using histogram and probability curves. From question https://groups.google.com/forum/#!topic/deap-users/Y8JX4AKwzhk
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| # Examples on how to extract and plot fitness diversity for a population, hall of fame, or | |
| # frontier using histogram and probability curves. Mostly working with multi objective fitness as per | |
| # question from https://groups.google.com/forum/#!topic/deap-users/Y8JX4AKwzhk | |
| # Derek M Tishler - 2019 | |
| import array | |
| import random | |
| import numpy as np | |
| import matplotlib as mpl | |
| mpl.use('Agg') # may need to remove this if using plt.show() | |
| import matplotlib.pyplot as plt | |
| from scipy import stats | |
| from deap import algorithms | |
| from deap import base | |
| from deap import creator | |
| from deap import tools | |
| def plot_diversity(hof): | |
| fig = plt.figure(figsize=(8,4), dpi=200) | |
| n_row = 1 | |
| n_col = len(hof[0].fitness.values) | |
| # Loop over the diff fitness dimensions and plot a histogram and probability curves | |
| for fit_tuple_idx in range(n_col): | |
| ax = plt.subplot2grid((n_row,n_col), (0, fit_tuple_idx), rowspan=1) | |
| # hof or paretofrontier is list of top or non-dominating front, either way its a list of individuals you can get fitness from | |
| #example: hof[0].fitness.values | |
| # so we can iteratare over the list and get the fitness, then plot histograms or any method you like to explore convergence | |
| fitnesses = [ind.fitness.values[fit_tuple_idx] for ind in hof] | |
| # lets plot a histogram of each fitness(or just the 1 if its not multi objective) | |
| n,bins,patches = ax.hist(fitnesses, histtype='stepfilled', bins=40, color='forestgreen' ,alpha=0.2, weights=np.ones_like(fitnesses)/float(len(fitnesses))) | |
| # To form a proba curve on the data i use numpy histogram for convenience and to ensure scaling for bin are readable for probabilities | |
| n, bin_edges = np.histogram(fitnesses, bins=40, density=True) | |
| norm_factor = 1./float(n.sum()) | |
| # now, also form a gauss kde on the DATA then scale to match the hist | |
| # note can increase the multipliers here and get a kde that falls to zero on endges, but its personal preff | |
| x_grid = np.linspace(np.min(fitnesses)-0.*np.std(fitnesses), np.max(fitnesses)+0.*np.std(fitnesses), 1000) | |
| kde_sample_evaluated = stats.gaussian_kde(fitnesses, bw_method=0.125).evaluate(x_grid) | |
| ax.plot(x_grid, kde_sample_evaluated*norm_factor, c='forestgreen', alpha=0.3, lw=0.6) | |
| ax.set_ylabel('probability') | |
| ax.set_xlabel(r'fitness$_{ %d}$'%(fit_tuple_idx+1)) | |
| ax.grid() | |
| plt.tight_layout() | |
| #plt.show(True) | |
| plt.savefig('hof_diversity.png') | |
| plt.close() | |
| def plot_scatter_w_diversity(hof, ext_ind_f=None, ext_ind_l=None): | |
| if ext_ind_f is not None: | |
| hof = [ext_ind_f] + hof | |
| if ext_ind_l is not None: | |
| hof = hof + [ext_ind_l] | |
| fig = plt.figure(figsize=(8,4), dpi=200) | |
| ax = plt.subplot2grid((1,2), (0, 0), rowspan=1) | |
| x = [float(ind.fitness.values[0]) for ind in hof] | |
| y = [float(ind.fitness.values[1]) for ind in hof] | |
| # labels to marry with figure in source whitepaper | |
| plt.text(x[0]-5., y[0]+1., "ext", fontsize=5, | |
| horizontalalignment='center', verticalalignment='center') | |
| plt.text(x[-1]-5., y[-1]-0.5, "ext", fontsize=5, | |
| horizontalalignment='center', verticalalignment='center') | |
| plt.text(x[0]+(x[0+1]-x[0])/2. - 3., y[0]+(y[0+1]-y[0])/2. + 2., r'd$_f$', fontsize=5, | |
| horizontalalignment='center', verticalalignment='center') | |
| plt.text(x[-1]+(x[-1]-x[-2])/2. - 3., y[-1]+(y[-2]-y[-1])/2. + 0., r'd$_l$', fontsize=5, | |
| horizontalalignment='center', verticalalignment='center') | |
| for i in range(1,len(x[1:-1])): | |
| plt.text(x[i]+(x[i+1]-x[i])/2. - 3., y[i]+(y[i+1]-y[i])/2. + 1., r'd$_{%d}$'%i, fontsize=5, | |
| horizontalalignment='center', verticalalignment='center') | |
| plt.scatter([x[0]],[y[0]], s=8, c='w', alpha=0.75, edgecolors='forestgreen', linewidth=1.0) | |
| plt.scatter([x[-1]],[y[-1]], s=8, c='w', alpha=0.75, edgecolors='forestgreen', linewidth=1.0) | |
| plt.scatter(x[1:-1],y[1:-1], s=8, c='forestgreen', alpha=0.75) | |
| plt.plot(x,y, lw=0.75, c='forestgreen', alpha=0.75) | |
| plt.grid() | |
| # https://www.iitk.ac.in/kangal/Deb_NSGA-II.pdf | |
| equation_1_page_188_str = r'$\Delta = \dfrac{d_f + d_l + \sum_{i=1}^{N-1} \left| d_i - \bar{d} \right|}{d_f + d_l + (N-1){\bar{d}}}$' | |
| def equation_1_page_188(hof): | |
| # assuming end points are extreme solutions(use a big pop for good data) | |
| d_l = np.linalg.norm(np.array(hof[-1].fitness.values)-np.array(hof[-2].fitness.values)) | |
| d_f = np.linalg.norm(np.array(hof[1].fitness.values)-np.array(hof[0].fitness.values)) | |
| N = float(len(hof)) | |
| d_i = np.abs([np.linalg.norm(np.array(ind.fitness.values)-np.array(hof[i+2].fitness.values)) for i,ind in enumerate(hof[1:-2])]) | |
| d_bar = np.mean(d_i) | |
| sum_ = np.sum([d-d_bar for d in d_i]) | |
| diversity_measure_del = (d_f + d_l + sum_)/(d_f + d_l + (N-1.)*d_bar) | |
| return diversity_measure_del | |
| diversity_measure_del = equation_1_page_188(hof) | |
| ax = plt.subplot2grid((1,2), (0, 1), rowspan=1) | |
| plt.text(0.5,0.7, equation_1_page_188_str, fontsize=17, | |
| horizontalalignment='center', verticalalignment='center', transform=ax.transAxes) | |
| plt.text(0.5,0.25, r"$\Delta = $%0.3f"%diversity_measure_del, fontsize=20, | |
| horizontalalignment='center', verticalalignment='center', transform=ax.transAxes) | |
| plt.axis('off') | |
| plt.tight_layout() | |
| #plt.show(True) | |
| if ext_ind_f is not None or ext_ind_l is not None: | |
| plt.savefig('hof_frontier_estimated_extreme.png') | |
| else: | |
| plt.savefig('hof_frontier.png') | |
| plt.close() | |
| ########## Can ignore this section, and replace with hof or pop list ########## | |
| # I create a fake pop with fitness values matchin the question's | |
| creator.create("FitnessMax", base.Fitness, weights=(1.0,1.0)) | |
| creator.create("Individual", array.array, typecode='b', fitness=creator.FitnessMax) | |
| toolbox = base.Toolbox() | |
| # Attribute generator | |
| toolbox.register("attr_bool", random.randint, 0, 1) | |
| # Structure initializers | |
| toolbox.register("individual", tools.initRepeat, creator.Individual, toolbox.attr_bool, 100) | |
| toolbox.register("population", tools.initRepeat, list, toolbox.individual) | |
| hof_fit_to_force = [ | |
| (173.384071,100.0), | |
| (45.082696,80.0), | |
| (34.562327,79.0), | |
| (24.557862,68.0), | |
| (14.550288,50.0), | |
| (13.726517,33.0), | |
| (13.479549,24.0), | |
| (13.439238,23.0), | |
| (13.430936,22.0), | |
| (13.409498,20.0), | |
| (9.029356,19.0), | |
| (7.88063,18.0), | |
| (2.611496,7.00), | |
| ] | |
| hof = toolbox.population(n=len(hof_fit_to_force)) | |
| for h,f in zip(hof, hof_fit_to_force): | |
| h.fitness.values = f | |
| # oversample original data | |
| #hof = toolbox.population(n=10000) | |
| #for h in hof: | |
| # h.fitness.values = [(x+((np.random.random()-0.5)*x),y+((np.random.random()-0.5)*y)) for x,y in [random.choice(hof_fit_to_force),]][0] | |
| # Done setting up for example, in your code you just use below and reference hof or pop | |
| ########################################################################### | |
| plot_diversity(hof) | |
| plot_scatter_w_diversity(hof) | |
| # if we have a method to est the extreme points, we can create 2 new inds and fill their fit with these estimates | |
| # lets split the hof in half, and build a std on each side to mimic asymetric distribution | |
| ext_sol_f = (hof[0].fitness.values[0]+np.std([ind.fitness.values[0] for ind in hof[:len(hof)/2]]), hof[0].fitness.values[1]+np.std([ind.fitness.values[1] for ind in hof[:len(hof)/2]])) | |
| ext_sol_l = (hof[-1].fitness.values[0]-np.std([ind.fitness.values[0] for ind in hof[-len(hof)/2:]]), hof[-1].fitness.values[1]-np.std([ind.fitness.values[1] for ind in hof[-len(hof)/2:]])) | |
| ext_ind_f = toolbox.individual() | |
| ext_ind_f.fitness.values = ext_sol_f | |
| ext_ind_l = toolbox.individual() | |
| ext_ind_l.fitness.values = ext_sol_l | |
| # override values will append to start and finish of hof. not very general but we know frontier shape? | |
| plot_scatter_w_diversity(hof, ext_ind_f=ext_ind_f, ext_ind_l=ext_ind_l) |
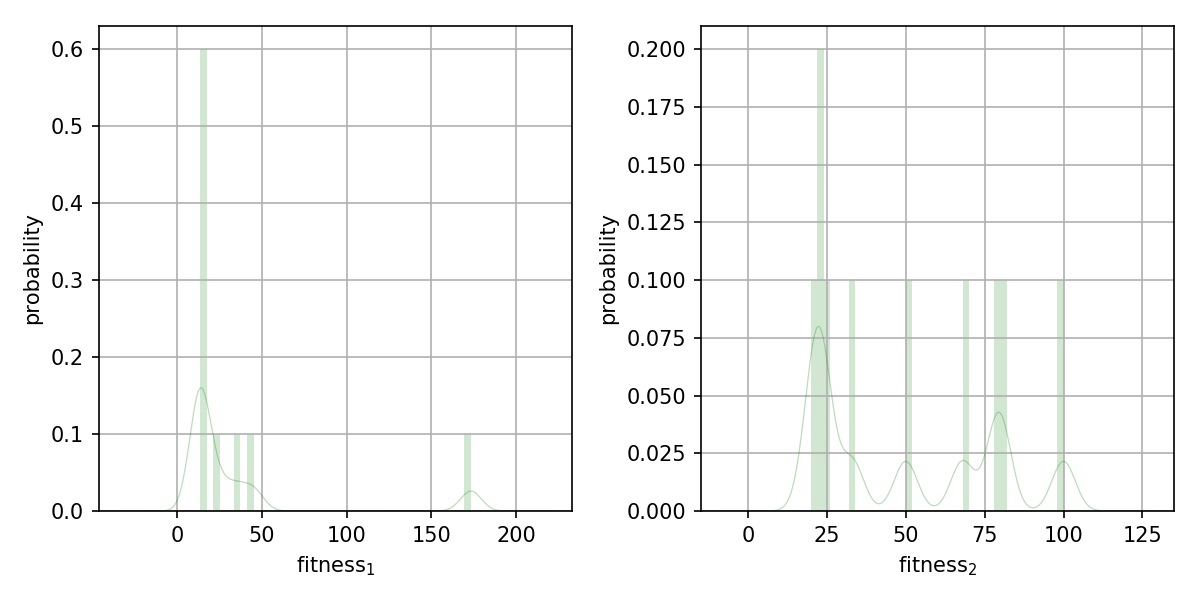
First chart is with a wider limit on the kde plot...with the default multiplier of 0. you would normally see the following figure:
So above is:
x_grid = np.linspace(np.min(fitnesses)-1.*np.std(fitnesses), np.max(fitnesses)+1.*np.std(fitnesses), 1000)
vs below's:
x_grid = np.linspace(np.min(fitnesses)-0.*np.std(fitnesses), np.max(fitnesses)+0.*np.std(fitnesses), 1000)

Now can attempt to measure diversity, without a benchmark, via diversity metric del found in paper:
A Fast and Elitist Multiobjective Genetic Algorithm: NSGA-II
Kalyanmoy Deb, Associate Member, IEEE, Amrit Pratap, Sameer Agarwal, and T. Meyarivan
https://www.iitk.ac.in/kangal/Deb_NSGA-II.pdf
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment




Resulting in a saved figure: