Generate the images like Yash 😃
System: Aalto Triton Linux On login node (CPU)
Since I met some problems when I run the workflow, I just used these parameters you provided in PP_Complex_Tip branch. The image I get is diffenrent from you. Images samples
Now, I am tring to go though the workflow.ipynb
I installed both OpenCL and complix_tip branch in diffent Conda environments, "PPComplexTip" and "PPOpenCL". PPComplexTip uses Python 3.8 and Numpy 19.5; PPOpenCL uses Python 2.7 And tested them using the Graphene examples. Both work well.
I install Runner according to https://github.com/SINGROUP/Runner, with just one minor change from numpy to numpy<=1.19.5 in setup.py as I found only Numpy lower than 1.19 can be used in PPAFM_Complex_tip.
Question 1:
# runner
pre_runner_data = RunnerData()
pre_runner_data.append_tasks('shell', 'module load anaconda')
pre_runner_data.append_tasks('shell', 'module load gcc')
runner = SlurmRunner('PPM',
pre_runner_data=pre_runner_data,
cycle_time=900,
max_jobs=50)
runner.to_database()TypeError Traceback (most recent call last) Cell In[15], line 5
3 pre_runner_data.append_tasks('shell', 'module load anaconda')
4 pre_runner_data.append_tasks('shell', 'module load gcc') ---->
5 runner = SlurmRunner('PPM',
6 pre_runner_data=pre_runner_data,
7 cycle_time=900,
8 max_jobs=50)
9 # runner = SlurmRunner('PPM',
10 # cycle_time=900,
11 # max_jobs=50)
12 runner.to_database() TypeError: __init__() got an unexpected keyword argument 'pre_runner_data'
It seems SlurmRunner do not have this parameter, pre_runner_data.
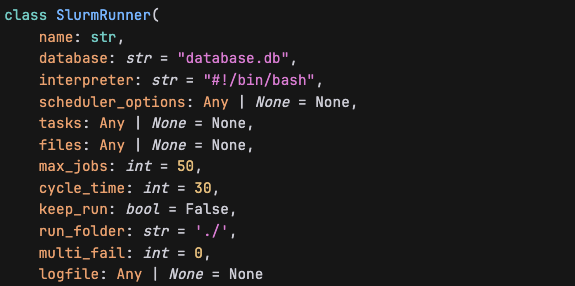
Question 2:
runner_data = RunnerData('get_ppm_data')
runner_data.scheduler_options = {'-n': 1,
'--time': '0-00:30:00',
'--mem-per-cpu': 12000}
runner_data.add_file('get_data.py')
runner_data.add_file('gen_params.py')
runner_data.add_file('run.sh', 'run_PPM.sh')
runner_data.append_tasks('python', 'gen_params.py', copy.deepcopy(params))
runner_data.append_tasks('shell', 'chmod +x prepare.sh')
runner_data.append_tasks('shell', 'chmod +x run_PPM.sh')
runner_data.append_tasks('shell', './prepare.sh')
runner_data.append_tasks('shell', './run_PPM.sh')
runner_data.append_tasks('python', 'get_data.py')
runner_data.append_tasks('shell', 'if [ -d PPM-complex_tip ]; then rm -rf PPM-complex_tip; fi')
runner_data.append_tasks('shell', 'if [ -d PPM-OpenCL ]; then rm -rf PPM-OpenCL; fi')
runner_data.keep_run = TrueError occurs when run this block directly. I should replace 'get_ppm_data' with some value. I gave an empty dictinary {}, it can prevent errors. Could you please tell me the right way here.
Question 4:
@interact(i=(1, 1), z=(40, 65))
def interactive_xy(i, z):
row = fdb.get(i)
np.save('test.npy', row.data['box'])
print(row.label, row.data['runner']['tasks'][0][2]['Amp'])
main('test.npy', z, [0.1], 2, 0, 0, None, None, 'A',
'dummy.png', _rotate=True)KeyError: 'box'
In my case, row.data seems doesn't have the attribute 'box'.
Then, the following blocks cannot running succesfully.
Question 5:
I noticed you used both PPAFM_Opencl and PPAFM_Complex_Tip. Without using this workflow, I just used the PPAFM_Complex_Tip. I thik this is a reason the images I obtained is not like you. I created two Conda environments on Triton to make two branches work. I wonder do I need to add some code to let this workflow know which environment to use?
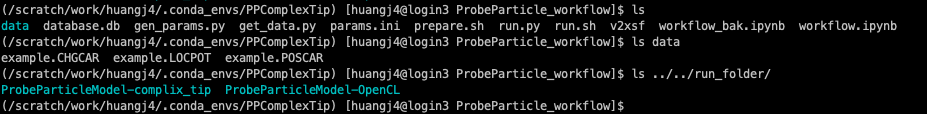
I created the folders according to some discribtions in prepare.sh
All the file used are stored here