This document describes a minor update to genomic SEM that provides the user with the option to control how the LD score intercept is used to apply genomic control to GenomicSEM GWAS and code to get quick initial genetic correlations and the standard errors of the genetic correlation from the ldsc() function.
Behind the scenes, and poorly documented (there were some comments in the code, that’s it), GenomicSEM was applying Genomic Control. The LD score regression intercept produces an expectation for the mean chi-square statistic under the null. As a chi2 distribution with 1 df has a mean of 1.0, an LDSC intercept greater than 1.0 can be used as an index of inflation of the test statistic attributable to uncontrolled confounding (Bulik Sullivan et al. 2015). Specifically, we estimate the univariate LD score intercept and inflate the SE of the estimated SNP-trait covariance by multiplying the SE by the intercept. This is a conservative procedure, and since the LD score intercept typically grows with growing sample sizes in GWAS, this procedure became increasingly conservative as all of us analyze larger and larger GWASs. We'll from now on inflate the standard error with sqrt(Intercept) and allow the user to change this behavior to the previous (conservative) default, or, turn of Genomic Control entirely (not something we would recommend you do unless you have a solid understanding of the nature of the LD score intercept and have good reason to ignore it).
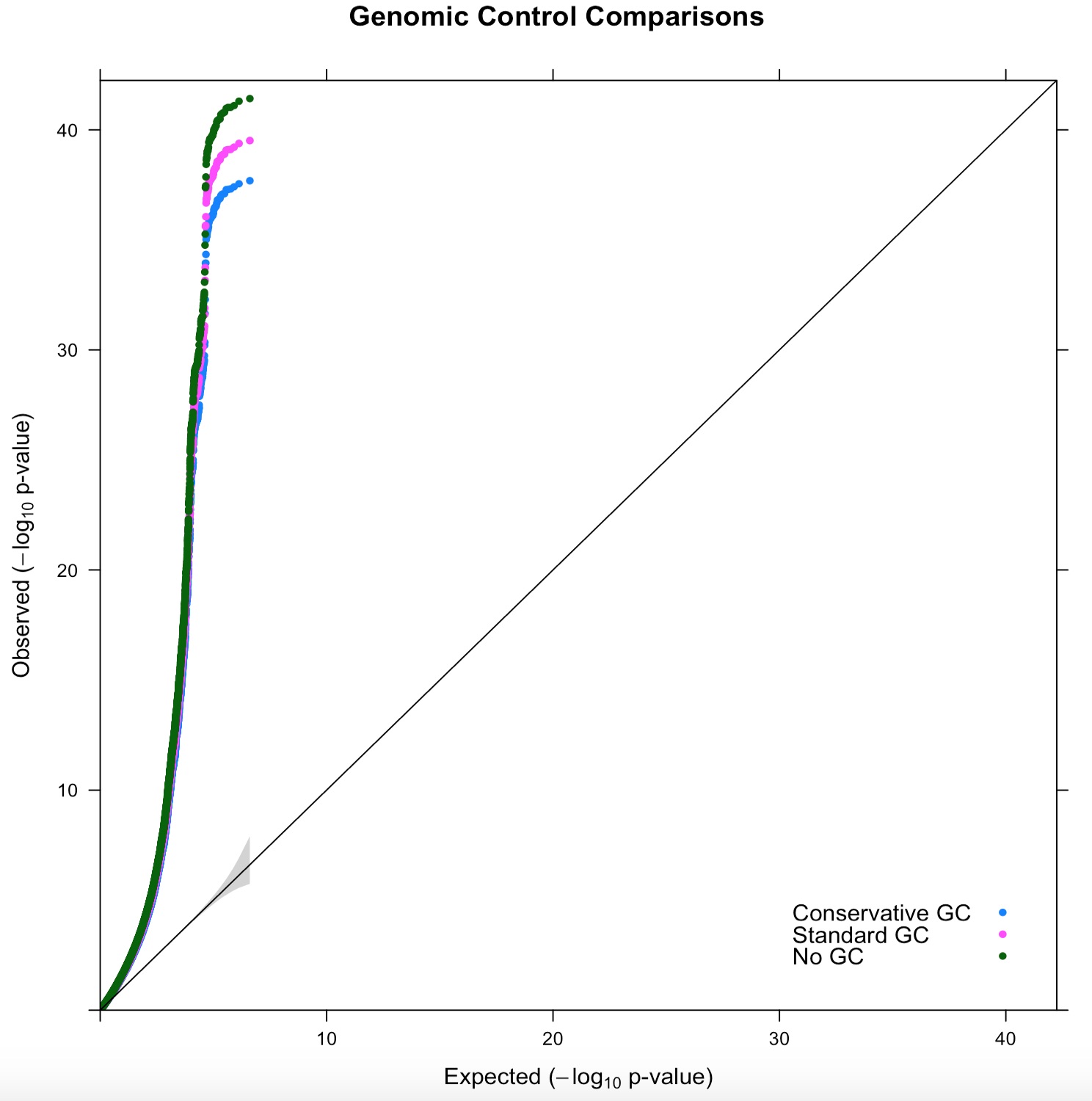
As an illustration we reran the single factor p-factor model first presented in Grotzinger et al. (2019) and inspect the effect on the QQ-plot, LD score intercept (and its s.e.), mean chi2 mean chi2 of the heterogeneity statistics (Q) and the significance of the SNP h2.
| Effective N | mean chi2 | mean chi2 (Q) | Univariate LDSC intercept | h2 Z-statistic | |
|---|---|---|---|---|---|
| No Genomic Control | 84,765 | 2.071 | 1.206 | 1.059 | 26.07 |
| Standard Genomic Control | 81,434 | 1.986 | 1.196 | 1.018 | 26.01 |
| Conservative Genomic Control | 78,173 | 1.906 | 1.186 | 0.980 | 25.89 |
You can also clearly spot some differences in the QQ-plots with: no, standard and conservative genomic control derived from a re-analysis in the p-factor model:
The user can set the argument GC = in the userGWAS() and commonfactorGWAS() to "none", "standard" (the default from now on) and "conserv" (the previous default). the previous default may produce different results from the previous versions of genomicSEM, because we continually push minor updates.
While its preferred, and easy enough, to estimate genetic correlation using a model supplied to the usermodel() function, we estimated a correlation matrix, and a matrix of the standard errors associated with it, in the output of the ldsc() function. set the argument stand= TRUE and ldsc() will produce thet standardized covariance matrix (i.e. the correlation matrix) and the sampling (co)variances of the elements in the correlation matrix. Though we emphasize the most accurate estimated of genetic correlations, and especially their standard errors are obtained by fitting a structural model, like the one below, in GenomicSEM:
cor.model <- '
lat1 =~ NA*trait1
lat2 =~ NA*trait2
trait1 ~~ 0*trait1 + 0*trait2
trait2 ~~ 0*trait2
lat1 ~~ 1*lat1
lat2 ~~ 1*lat2
lat1 ~~ cor*lat2
'
This model can be fit with usermodel() and it estimates the correlation between variables trait1 and trait2 as the parameter cor.