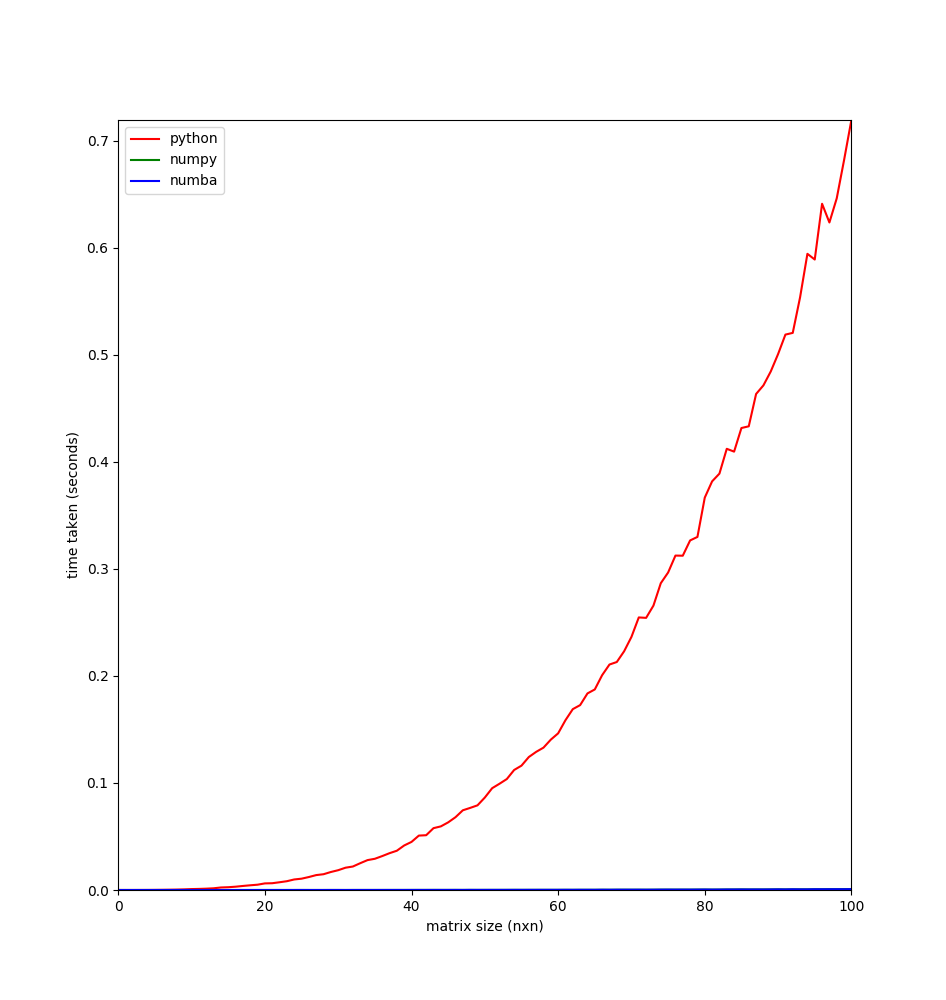
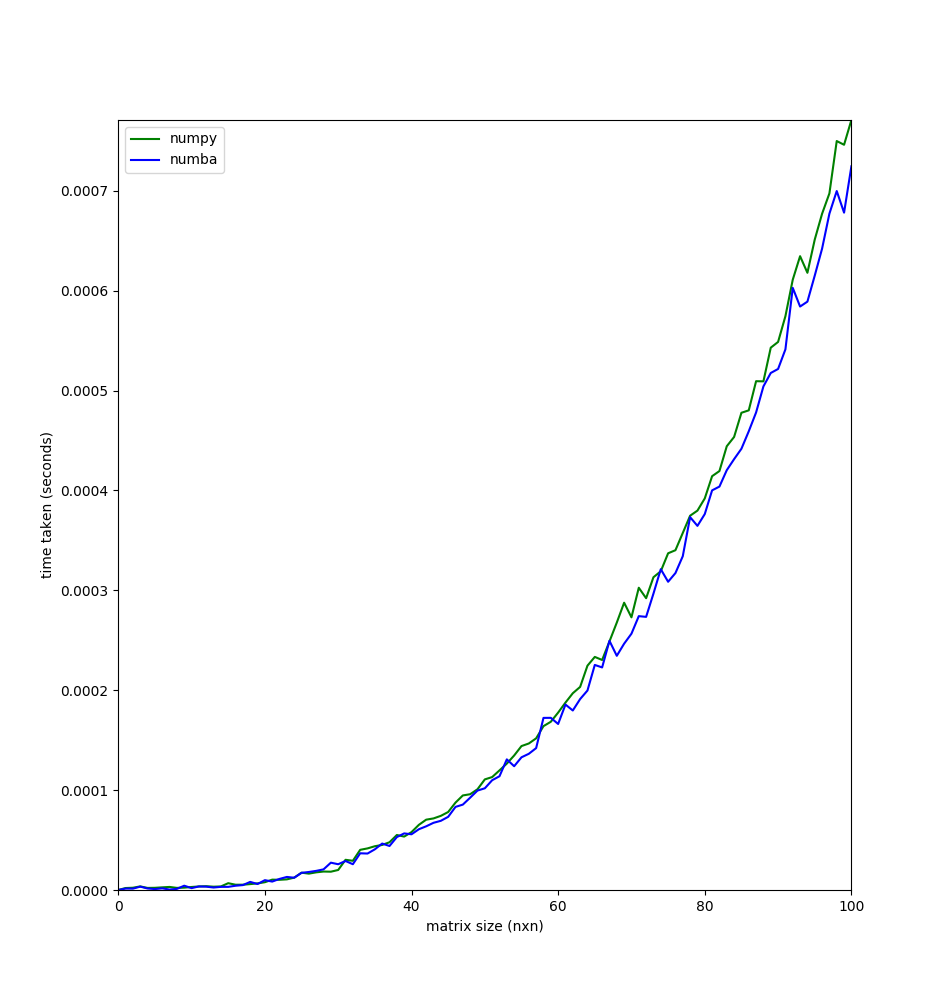
|
import random |
|
import time |
|
import timeit |
|
import argparse |
|
import sys |
|
|
|
import matplotlib.pyplot as plt |
|
import numba |
|
import numba.typed |
|
import numpy as np |
|
|
|
MIN = 0 |
|
MAX = 100 |
|
|
|
|
|
def gen_rand_mat(n): |
|
return np.random.randint(MIN, MAX, size=(n, n)) |
|
|
|
|
|
def mat_mul(A, B): |
|
C = np.zeros((A.shape[0], B.shape[1]), dtype=A.dtype) |
|
|
|
for r in range(A.shape[0]): |
|
for c in range(B.shape[1]): |
|
for i in range(len(A[0])): |
|
C[r, c] += A[r, i] * B[i, c] |
|
|
|
return C |
|
|
|
|
|
@numba.jit(nopython=True) |
|
def nu_mat_mul(A, B): |
|
C = np.zeros((A.shape[0], B.shape[1]), dtype=A.dtype) |
|
|
|
for r in range(A.shape[0]): |
|
for c in range(B.shape[1]): |
|
for i in range(len(A[0])): |
|
C[r, c] += A[r, i] * B[i, c] |
|
|
|
return C |
|
|
|
|
|
def main(argv): |
|
parser = argparse.ArgumentParser() |
|
parser.add_argument('max_n', nargs='?', default=10, type=int) |
|
parser.add_argument('--no-python', action='store_true') |
|
parser.add_argument('--no-numpy', action='store_true') |
|
parser.add_argument('--no-numba', action='store_true') |
|
parser.add_argument('--trials', type=int, default=1000, |
|
help='number of trials to take average of for each matrix size') |
|
|
|
args = parser.parse_args(argv[1:]) |
|
|
|
plt.ion() |
|
|
|
fig = plt.figure() |
|
ax = fig.add_subplot(111) |
|
|
|
ax.set_xlabel('matrix size (nxn)') |
|
ax.set_ylabel('time taken (seconds)') |
|
|
|
x = [0] |
|
|
|
lines = [] |
|
legends = [] |
|
|
|
if not args.no_python: |
|
py_times = [0] |
|
py_line = ax.plot(x, py_times, "r")[0] |
|
lines.append(py_line) |
|
legends.append('python') |
|
|
|
if not args.no_numpy: |
|
np_times = [0] |
|
np_line = ax.plot(x, np_times, "g")[0] |
|
ax.legend([np_line], ['numpy']) |
|
lines.append(np_line) |
|
legends.append('numpy') |
|
|
|
if not args.no_numba: |
|
nu_times = [0] |
|
nu_line = ax.plot(x, nu_times, "b")[0] |
|
ax.legend([nu_line], ['numba']) |
|
lines.append(nu_line) |
|
legends.append('numba') |
|
|
|
ax.legend(lines, legends) |
|
|
|
nu_mat_mul(gen_rand_mat(1), gen_rand_mat(1)) |
|
|
|
for i in range(1, args.max_n + 1): |
|
x.append(i) |
|
|
|
gen_time = timeit.timeit("gen_rand_mat({})".format(i), number=args.trials * 2, globals=globals()) |
|
m = 0 |
|
|
|
if not args.no_python: |
|
py_statement = "mat_mul(gen_rand_mat({0}), gen_rand_mat({0}))".format(i, i) |
|
py_times.append((timeit.timeit(py_statement, number=args.trials, globals=globals()) - gen_time) / args.trials) |
|
py_line.set_xdata(x) |
|
py_line.set_ydata(py_times) |
|
m = max(m, *py_times) |
|
|
|
if not args.no_numpy: |
|
np_statement = "np.dot(gen_rand_mat({0}), gen_rand_mat({0}))".format(i, i) |
|
np_times.append((timeit.timeit(np_statement, number=args.trials, globals=globals()) - gen_time) / args.trials) |
|
np_line.set_xdata(x) |
|
np_line.set_ydata(np_times) |
|
m = max(m, *np_times) |
|
|
|
if not args.no_numba: |
|
nu_statement = "nu_mat_mul(gen_rand_mat({0}), gen_rand_mat({0}))".format(i, i) |
|
nu_times.append((timeit.timeit(nu_statement, number=args.trials, globals=globals()) - gen_time) / args.trials) |
|
nu_line.set_xdata(x) |
|
nu_line.set_ydata(nu_times) |
|
m = max(m, *nu_times) |
|
|
|
ax.set_xlim(0, i) |
|
ax.set_ylim(0, m) |
|
|
|
fig.canvas.draw() |
|
fig.canvas.flush_events() |
|
|
|
plt.ioff() |
|
plt.show() |
|
|
|
|
|
if __name__ == "__main__": |
|
main(sys.argv) |