-
-
Save Svidro/8f9c06e2c8bcae214cdd7aa9afe57c50 to your computer and use it in GitHub Desktop.
| Scripts mostly taken from Pete, and also from the forums. For easy access and reference. | |
| Covers both Selecting, which is necessary to run most commands like cell detection, and collecting objects into a variable | |
| for processing. The latter is more efficient when you do not need to run a command like cell detection. | |
| TOC | |
| A Selection guide.groovy - not actually a script, just a collection of useful tips. | |
| Access top level objects.groovy - Creates a list of all objects at the "top" of the hierarchy. This list is dynamic. | |
| Decendant and child objects.groovy - Shows accessing child objects or decendant objects. | |
| Get objects within another object.groovy - ignores hierarchy in case of overlaps! | |
| SelectClassificationsByName.groovy - Select objects based on their classifications having certain words | |
| Selecting objects within a drawn annotation.groovy - <- What it says. Removes the drawn/selected annotation. | |
| Selecting objects touching an annotation.groovy - uses Geometry functions to determine if the edges of objects are touching. | |
| Testing selection models.groovy - For my reference. | |
| Using selectionModel for groups of objects.groovy - Very basic example of creating a selection based on some thresholding. |
| //Warning, some functions like getArea() will not work on some types of objects, like points, and will cause the script to crash | |
| //if you are searching across both area based annotations and points. getArea() also returns measurements IN PIXELS. | |
| //Select all of a type | |
| selectAnnotations(); | |
| selectDetections(); | |
| //get as complicated as you want in the selection here, goes through all objects | |
| selectObjects { p -> p.getPathClass() == getPathClass("Stroma") && p.isAnnotation() } | |
| //or from https://groups.google.com/forum/#!topic/qupath-users/PxrCwx5ttXI | |
| selectObjects { | |
| //Some criteria here | |
| return it.isAnnotation() && it.getPathClass() == getPathClass('Tumor') | |
| } | |
| //These do not select, but each can be used to generate a list of objects, which can then be used... | |
| getAnnotationObjects() | |
| getDetectionObjects() | |
| getSelectedObject() | |
| getSelectedObjects() | |
| //... in this type of structure | |
| def Annotations = getAnnotationObjects() | |
| getCurrentHierarchy().getSelectionModel().setSelectedObjects(Annotations, null) | |
| //The total of these two lines is the same as selectAnnotations(), but is capable of being modified by: | |
| def smallAnnotations = getAnnotationObjects().findAll {it.getROI().getArea() < 400000} | |
| //similar to the selectObjects code above | |
| //The 400000 in this case is in pixels, and would need to be modified by a metadata call (which varies depending on your version of QuPath) | |
| //Selecting multiple single object in a row | |
| resetSelection() | |
| getCellObjects().each{ | |
| getCurrentHierarchy().getSelectionModel().setSelectedObject(it, true); | |
| threshold = measurement(it, "Nucleus: Channel 2 mean")+measurement(it, "Nucleus: Channel 2 std dev") | |
| runPlugin('qupath.imagej.detect.cells.SubcellularDetection', '{"detection[Channel 1]": -1.0, "detection[Channel 2]": '+threshold+', "detection[Channel 3]": -1.0, "doSmoothing": true, "splitByIntensity": false, "splitByShape": true, "spotSizeMicrons": 1.0, "minSpotSizeMicrons": 1, "maxSpotSizeMicrons": 2.0, "includeClusters": true}'); | |
| resetSelection(); | |
| fireHierarchyUpdate() | |
| } | |
| //In 0.2.0 many things can be replaced by simple thresholding and then using scripts like: | |
| selectObjectsByClassification("Islet"); | |
| selectObjectsByClassification("Islet", "Stroma"); | |
| //Which directly selects objects in one line. | |
| //Selecting objects within other objects, that may not match up with the hierarchy | |
| //https://forum.image.sc/t/qupath-getting-detectionsobjects-within-non-parent-annotation/41784/2 | |
| def hierarchy = getCurrentHierarchy() | |
| def parent = getSelectedObject() | |
| def objects = hierarchy.getObjectsForROI(null, parent.getROI()) | |
| .findAll { it.isDetection() } | |
| hierarchy.getObjectsForROI(qupath.lib.objects.PathDetectionObject, parent.getROI()) |
| //accesses the highest level objects without cycling through all objects or annotations | |
| //Important to note that this list is DYNAMIC and will adjust as objects are created or destroyed. | |
| //0.1.2 or 0.2.0 | |
| //may want to resolveHierarchy() before running this in 0.2.0 | |
| topLevel = getCurrentHierarchy().getRootObject().getChildObjects() |
| //Finding decendant and child objects of annotations | |
| //0.1.2 | |
| // Define classes | |
| class1 = getPathClass('class1') | |
| class2 = getPathClass('class2') | |
| // Loop through and add child/descendant counts | |
| hierarchy = getCurrentHierarchy() | |
| getAnnotationObjects().each { | |
| if (it.getPathClass() != class1) | |
| return false | |
| def children = it.getChildObjects() | |
| def nClass2Children = children.count {it.getPathClass() == class2} | |
| def descendants = hierarchy.getDescendantObjects(it, null, null) | |
| def nClass2Descendants = descendants.count {it.getPathClass() == class2} | |
| it.getMeasurementList().putMeasurement('Number of ' + class2 + ' child objects', nClass2Children) | |
| it.getMeasurementList().putMeasurement('Number of ' + class2 + ' descendant objects', nClass2Descendants) | |
| it.getMeasurementList().closeList() | |
| } |
| //Sara McArdle | |
| // 0.2.0 assigns a pathClass to detections that have an exactly matching annotation. | |
| import static qupath.lib.gui.scripting.QPEx.* | |
| def annots=getAnnotationObjects() | |
| def annotgeos=annots.collect{it.getROI().getGeometry()} | |
| def dets=getDetectionObjects() | |
| def detgeos=dets.collect{it.getROI().getGeometry()} | |
| annotgeos.eachWithIndex{a, idx-> | |
| def equality=detgeos.collect{it.equalsTopo(a)} | |
| int match=equality.findIndexOf {it==true} | |
| if (match>=0){ | |
| dets[match].setPathClass(annots[idx].getPathClass()) | |
| } | |
| } |
| //0.2.0 only option for ignoring the hierarchy and selecting objects within another object. | |
| //https://forum.image.sc/t/qupath-getting-detectionsobjects-within-non-parent-annotation/41784/2 | |
| //Stores the objects you want within a variable called "objects" | |
| //Original: | |
| def hierarchy = getCurrentHierarchy() | |
| def parent = getSelectedObject() | |
| def objects = hierarchy.getObjectsForROI(null, parent.getROI()) | |
| .findAll { it.isDetection() } | |
| //one line version | |
| def objects = getCurrentHierarchy().getObjectsForROI(qupath.lib.objects.PathDetectionObject, getSelectedObject().getROI()) | |
| //If you have single Annotations with different classifications, you could use something like the following | |
| //Example, you have a Tissue annotation, and within that a specific Tumor annotation. You want the "cells" within the Tumor annotation | |
| //[0] at the end of the next line gets the first (and only, in this example) annotation that is classified as Tumor. | |
| tumorAnno = getAnnotationObjects().findAll{it.getPathClass() == getPathClass("Tumor")}[0] | |
| objects = getCurrentHierarchy().getObjectsForROI(qupath.lib.objects.PathCellObject, tumorAnno.getROI()) |
| /** | |
| * Source https://gist.github.com/petebankhead/abb3f5a3a2b55ddc22f709c99964857e | |
| * Helper script for QuPath to find objects with one or more classifications. | |
| * | |
| Mostly replaced by | |
| selectObjectsByClassification("Islet"); | |
| in 0.2.0 | |
| * @author Pete Bankhead | |
| */ | |
| // Insert as many or few classifications here as required | |
| selectObjects {checkForClassifications(it.getPathClass(), 'CD8', 'FoxP3')} | |
| print 'Selected ' + getSelectedObjects().size() | |
| boolean checkForSingleClassification(def pathClass, classificationName) { | |
| if (pathClass == null) | |
| return false | |
| if (pathClass.getName() == classificationName) | |
| return true | |
| return checkForSingleClassification(pathClass.getParentClass(), classificationName) | |
| } | |
| boolean checkForClassifications(def pathClass, String...classificationNames) { | |
| if (classificationNames.length == 0) | |
| return false | |
| for (String name : classificationNames) { | |
| if (!checkForSingleClassification(pathClass, name)) | |
| return false | |
| } | |
| return true | |
| } |
| //Sara McCardle | |
| import static qupath.lib.gui.scripting.QPEx.* | |
| def islets=getDetectionObjects() | |
| def isletsgeos=islets.collect{it.getROI().getGeometry()} | |
| def outsidegeo=getAnnotationObjects().find{it.getPathClass()==getPathClass("Tissue")}.getROI().getGeometry() | |
| def intersections=[] | |
| isletsgeos.eachWithIndex{entry,idx-> | |
| if (entry.intersects(outsidegeo)){ | |
| intersections<<idx | |
| } | |
| } | |
| print(intersections) | |
| getCurrentHierarchy().getSelectionModel().selectObjects(islets[intersections]) |
| //This script allows you to draw a quick annotation, and then select all objects within it for... whatever reason | |
| //It also deletes the drawn annotation by design with "clearSelectedObjects()" so comment out that line if you | |
| //want to keep your annotation | |
| // 0.1.2 | |
| // Get the current selected object & hierarchy | |
| selected = getSelectedObject() | |
| hierarchy = getCurrentHierarchy() | |
| // Get all the objects inside the current selection | |
| objectsToSelect = hierarchy.getDescendantObjects(selected, null, null) | |
| clearSelectedObjects() | |
| if (objectsToSelect != null) { | |
| // Remove the current selected object | |
| hierarchy.removeObject(selected, true) | |
| // Update the selection | |
| hierarchy.getSelectionModel().selectObjects(objectsToSelect) | |
| } |
| //This is just a set of tests for figuring out how selection models work. | |
| //0.1.2 | |
| //Select Tumor annotations within named Organ annotations | |
| hierarchy = getCurrentHierarchy() | |
| hierarchy.getSelectionModel().clearSelection(); | |
| def organs = getAnnotationObjects().findAll {it.getDisplayedName().equalsIgnoreCase("Organ")} | |
| organs.each{hierarchy.getSelectionModel().selectObjects(hierarchy.getDescendantObjects(it, null, null).findAll{it.getPathClass() == getPathClass("Tumor")})} | |
| //Another variant | |
| def hierarchy = getCurrentHierarchy() | |
| hierarchy.getSelectionModel().clearSelection() | |
| hierarchy.getSelectionModel().selectObjects(getAnnotationObjects().findAll {it.getPathClass() == getPathClass("Tumor") && | |
| it.getParent().getDisplayedName().equalsIgnoreCase("Organ") && it.getLevel() != 1}) |
| //This script is a modification of the script to find annotations of a certain size, which then uses | |
| //getSelectionModel to take the list of annotations and have QuPath register them as selected | |
| //getArea() returns a measurement in PIXELS not square microns. | |
| //0.1.2 0.2.0 | |
| def smallAnnotations = getAnnotationObjects().findAll {it.getROI().getArea() < 400000} | |
| //This line does the selecting, and you should be able to swap in any list of objects for smallAnnotations | |
| getCurrentHierarchy().getSelectionModel().setSelectedObjects(smallAnnotations, null) |
@simonesteffens Ah, just as a warning, this is not the best place to post since there are no warnings for Gists! I just happened to be checking this 10 hours later... but it could have been weeks!
That said, I am not entirely sure what you are trying to do again, but I think your top arrow is actually pointing to the total number of "objects" of that class. I am someone confused as to whether you have SLIC detections and SLICs converted into cells at the same time, but you should be able to apply any measurement to the parent annotation.
Code would go something like:
annotations = getAnnotationObjects();
annotations.each{
annoClass = it.getPathClass();
if(annoClass){
detections = it.getChildObjects().findAll{d-> d.isDetection()}
//This line adds a new measurement, the class name, along with the number of detections, to the parent annotation
it.getMeasurementList().putMeasurement(annoClass.getName(), detections .size())
//This line makes every object have the same class as its parent
detections.each{it.setPathClass(annoClass)}
}
}
That is assuming that you do have detection objects as children to the annotations. With the amount of yellow showing up in your screenshots, I think something else might be going on. I suspect your hierarchy looks a bit different, and I am not really sure which script you used... there are a lot of them! Also, using the above script, you shouldn't need to convert the SLICs into cells, it is just looking for detections.
If you want an actual cell count, running cell detection will delete all of the SLIC detections, and you could use the above script to add cell counts/classify cells.
And another option, if you have cells, would be to use the one liner setCellIntensityClassifications("Nucleus: Area", 0) to make all cells everywhere positive. Then you would have a number of positive cells in your tumor and stroma annotations.
Hey, thanks for your answer!!!
It helped me a lot to change every area separately in cells.
I have one more question, maybe you will understand it better with the pics.
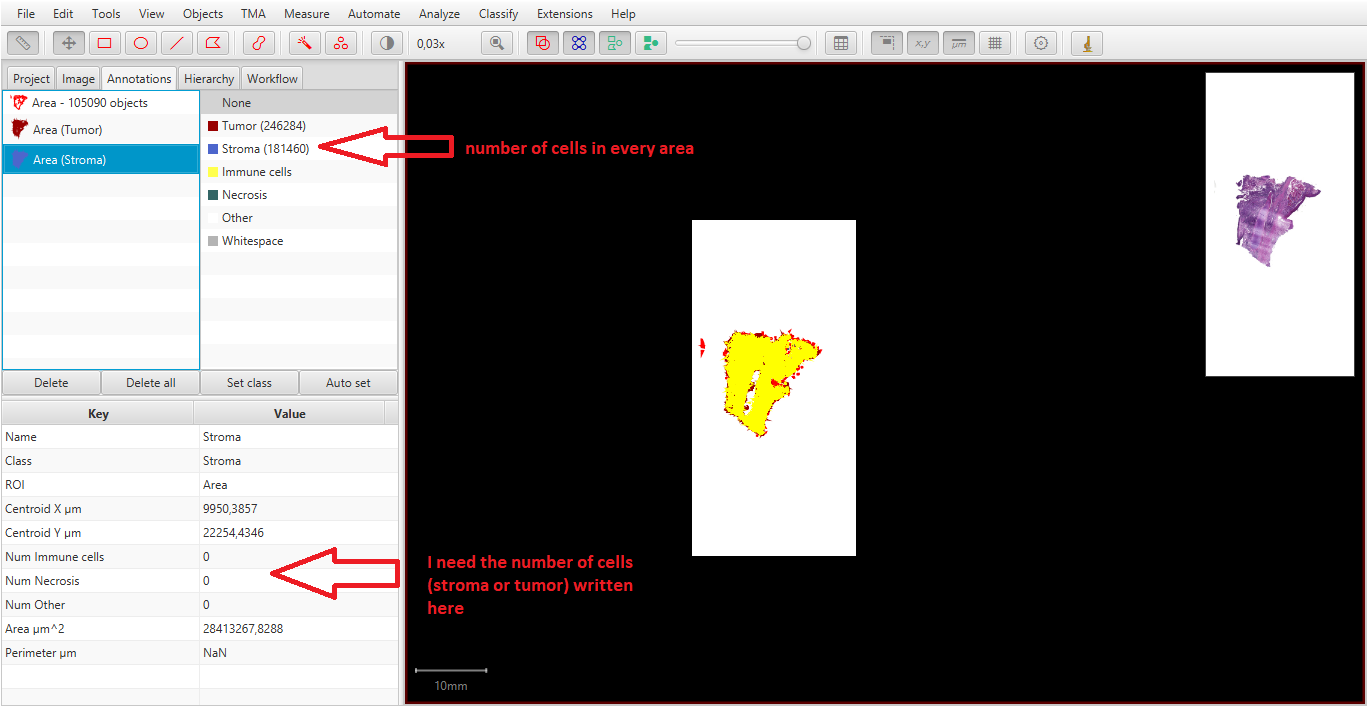
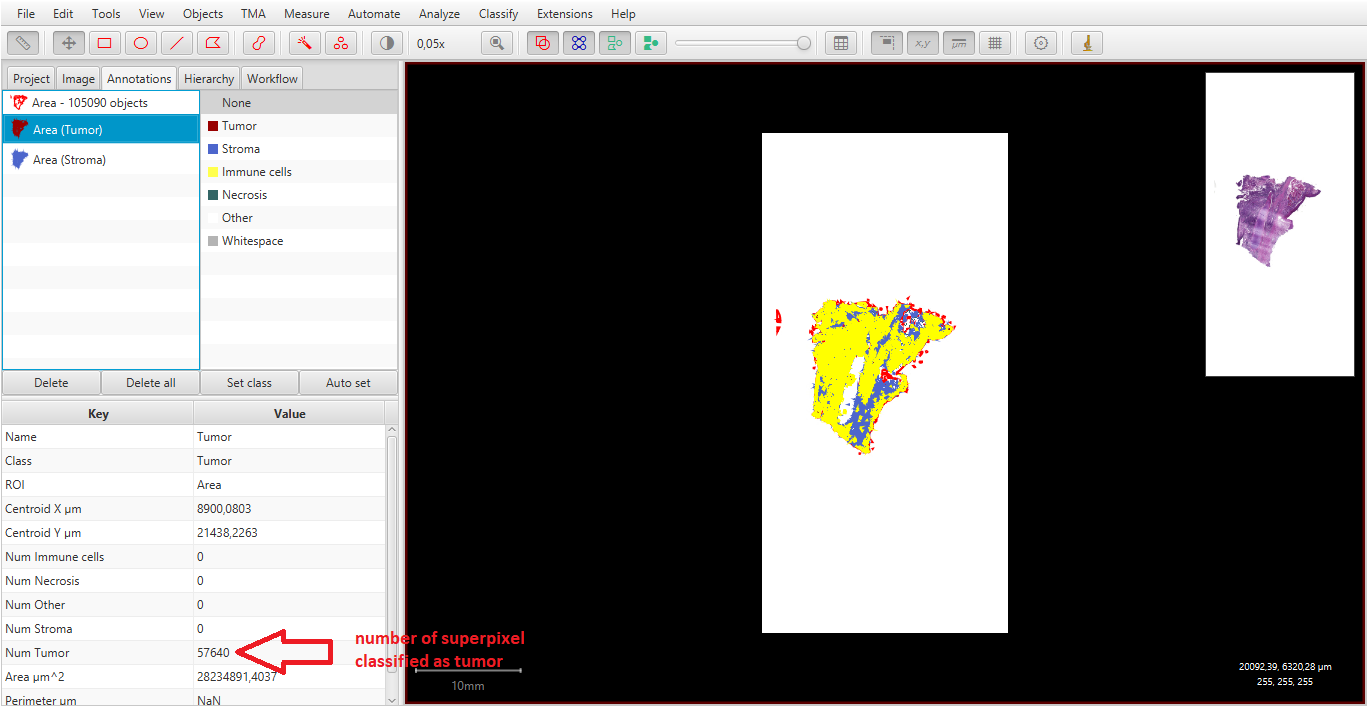
I changed in every area (Tumor and stroma) all superpixels to cells by selecting the area and running cell detection (with your script).
I got a result ( first picture), wich I can´t save by a script in a special document (xls). I should have the number of cells as annotation measurements (second picture) to run a script for saving. I think it´s because the cells are part of the area but the aren´t classified.
So is there a possibility to classify all cells in one annotation (area) as the same class? Without training the classifier? Or is it possible to get the number of cells of every annotation as annotation measurement without classifing again? Hope you did understand what I meant.
Thanks for your help :)