Created
January 11, 2017 11:22
-
-
Save aaronsaunders/6960eebecb7a9bc0999e02e08d79ed0b to your computer and use it in GitHub Desktop.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| { | |
| "cells": [ | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "# UPARSE analysis pipeline form amplicon sequences\n", | |
| "\n", | |
| "We will follow the Illumina paired end tutorial.\n", | |
| "\n", | |
| "http://drive5.com/usearch/manual/upp_ill_pe.html\n", | |
| "\n", | |
| "\n", | |
| "This page gives an example of 2 pipelines using UPARSE for Illumina paired reads. Both use the same preprocessing.\n", | |
| "\n", | |
| "### Preprocessing\n", | |
| "\n", | |
| "These commands make the following assumptions (which are usually but not always true).\n", | |
| "\n", | |
| "\n", | |
| "1. The forward and reverse read have a significant overlap, say 32 bases or more.\n", | |
| "\n", | |
| "2. There are no non-biological bases in the read such as adapters or barcodes.\n", | |
| "\n", | |
| "3. Sequences complementary to PCR primers are not included in the reads.\n", | |
| "\n", | |
| "4. The reads have been demultiplexed, i.e. split into separate FASTQ files for each sample.\n", | |
| "\n", | |
| "5. The FASTQ filenames start with the sample name.\n", | |
| "\n", | |
| "6. The reads are all on the same strand.\n", | |
| "\n", | |
| "\n", | |
| "Commands:\n", | |
| "\n", | |
| " usearch -fastq_mergepairs *_R1_*.fastq -relabel @ -fastqout merged.fq\n", | |
| "\n", | |
| " usearch -fastq_filter merged.fq -fastq_maxee 1.0 -relabel Filt -fastaout filtered.fa\n", | |
| "\n", | |
| " usearch -fastx_uniques filtered.fa -relabel Uniq -sizeout -fastaout uniques.fa\n", | |
| "\n", | |
| "\n", | |
| "### Original UPARSE (with UCHIME)\n", | |
| "\n", | |
| "http://drive5.com/usearch/manual/uparse_pipeline.html\n", | |
| "\n", | |
| "Takes the good quality, unique reads and clusters at 97% to identify centroids (which are screened for chimeras with UCHIME). Then clusters all of the sequences within 3% of a centroid as an 97% OTU.\n", | |
| "\n", | |
| "\n", | |
| "Commands:\n", | |
| "\n", | |
| " usearch -cluster_otus uniques.fa -minsize 2 -otus otus.fa -relabel Otu\n", | |
| "\n", | |
| " usearch -usearch_global merged.fq -db otus.fa -strand plus -id 0.97 \\\n", | |
| " -otutabout otutab.txt -biomout otutab.json\n", | |
| "\n", | |
| " \n", | |
| " ### UNOISE pipeline (NEW!)\n", | |
| " http://www.drive5.com/usearch/manual/unoise_pipeline.html\n", | |
| " \n", | |
| "Takes the good quality, unique reads and identifies centroids with an error model. These are also screened for chimeras (with UCHIME) in the same script. Then clusters all of the sequences within 3% of a centroid as an 97% OTU.\n", | |
| "\n", | |
| "Commands:\n", | |
| " \n", | |
| " usearch -unoise uniques.fa -tabbedout out.txt -fastaout denoised.fa\n", | |
| "\n", | |
| " usearch -usearch_global reads.fq -db denoised.fa -strand plus -id 0.97 -otutabout otu_table.txt" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "# Download Sample data" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 1, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "!rm -rf data # delete output and start fresh\n", | |
| "!mkdir data" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 2, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "fetch data\n", | |
| "URL transformed to HTTPS due to an HSTS policy\n", | |
| "--2017-01-11 10:20:31-- https://www.mothur.org/w/images/d/d6/MiSeqSOPData.zip\n", | |
| "Resolving www.mothur.org (www.mothur.org)... 141.214.120.22\n", | |
| "Connecting to www.mothur.org (www.mothur.org)|141.214.120.22|:443... connected.\n", | |
| "HTTP request sent, awaiting response... 200 OK\n", | |
| "Length: 36910055 (35M) [application/zip]\n", | |
| "Saving to: ‘data/MiSeqSOPData.zip’\n", | |
| "\n", | |
| "MiSeqSOPData.zip 100%[===================>] 35.20M 598KB/s in 86s \n", | |
| "\n", | |
| "2017-01-11 10:21:58 (419 KB/s) - ‘data/MiSeqSOPData.zip’ saved [36910055/36910055]\n", | |
| "\n", | |
| "Archive: data/MiSeqSOPData.zip\n", | |
| " creating: MiSeq_SOP/\n", | |
| " inflating: MiSeq_SOP/F3D0_S188_L001_R1_001.fastq \n", | |
| " creating: __MACOSX/\n", | |
| " creating: __MACOSX/MiSeq_SOP/\n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D0_S188_L001_R1_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D0_S188_L001_R2_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D0_S188_L001_R2_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D141_S207_L001_R1_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D141_S207_L001_R1_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D141_S207_L001_R2_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D141_S207_L001_R2_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D142_S208_L001_R1_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D142_S208_L001_R1_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D142_S208_L001_R2_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D142_S208_L001_R2_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D143_S209_L001_R1_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D143_S209_L001_R1_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D143_S209_L001_R2_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D143_S209_L001_R2_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D144_S210_L001_R1_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D144_S210_L001_R1_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D144_S210_L001_R2_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D144_S210_L001_R2_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D145_S211_L001_R1_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D145_S211_L001_R1_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D145_S211_L001_R2_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D145_S211_L001_R2_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D146_S212_L001_R1_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D146_S212_L001_R1_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D146_S212_L001_R2_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D146_S212_L001_R2_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D147_S213_L001_R1_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D147_S213_L001_R1_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D147_S213_L001_R2_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D147_S213_L001_R2_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D148_S214_L001_R1_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D148_S214_L001_R1_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D148_S214_L001_R2_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D148_S214_L001_R2_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D149_S215_L001_R1_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D149_S215_L001_R1_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D149_S215_L001_R2_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D149_S215_L001_R2_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D150_S216_L001_R1_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D150_S216_L001_R1_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D150_S216_L001_R2_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D150_S216_L001_R2_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D1_S189_L001_R1_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D1_S189_L001_R1_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D1_S189_L001_R2_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D1_S189_L001_R2_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D2_S190_L001_R1_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D2_S190_L001_R1_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D2_S190_L001_R2_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D2_S190_L001_R2_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D3_S191_L001_R1_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D3_S191_L001_R1_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D3_S191_L001_R2_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D3_S191_L001_R2_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D5_S193_L001_R1_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D5_S193_L001_R1_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D5_S193_L001_R2_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D5_S193_L001_R2_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D6_S194_L001_R1_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D6_S194_L001_R1_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D6_S194_L001_R2_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D6_S194_L001_R2_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D7_S195_L001_R1_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D7_S195_L001_R1_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D7_S195_L001_R2_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D7_S195_L001_R2_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D8_S196_L001_R1_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D8_S196_L001_R1_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D8_S196_L001_R2_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D8_S196_L001_R2_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D9_S197_L001_R1_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D9_S197_L001_R1_001.fastq \n", | |
| " inflating: MiSeq_SOP/F3D9_S197_L001_R2_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._F3D9_S197_L001_R2_001.fastq \n", | |
| " inflating: MiSeq_SOP/HMP_MOCK.v35.fasta \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._HMP_MOCK.v35.fasta \n", | |
| " inflating: MiSeq_SOP/Mock_S280_L001_R1_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._Mock_S280_L001_R1_001.fastq \n", | |
| " inflating: MiSeq_SOP/Mock_S280_L001_R2_001.fastq \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._Mock_S280_L001_R2_001.fastq \n", | |
| " inflating: MiSeq_SOP/mouse.dpw.metadata \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._mouse.dpw.metadata \n", | |
| " inflating: MiSeq_SOP/mouse.time.design \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._mouse.time.design \n", | |
| " inflating: MiSeq_SOP/stability.batch \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._stability.batch \n", | |
| " inflating: MiSeq_SOP/stability.files \n", | |
| " inflating: __MACOSX/MiSeq_SOP/._stability.files \n", | |
| "Fetch taxonomy reference data\n", | |
| "--2017-01-11 10:22:00-- http://drive5.com/sintax/rdp_16s_v16.fa.gz\n", | |
| "Resolving drive5.com (drive5.com)... 199.195.116.69\n", | |
| "Connecting to drive5.com (drive5.com)|199.195.116.69|:80... connected.\n", | |
| "HTTP request sent, awaiting response... 200 OK\n", | |
| "Length: 4602323 (4.4M) [application/x-gzip]\n", | |
| "Saving to: ‘data/rdp_16s_v16.fa.gz’\n", | |
| "\n", | |
| "rdp_16s_v16.fa.gz 100%[===================>] 4.39M 2.44MB/s in 1.8s \n", | |
| "\n", | |
| "2017-01-11 10:22:03 (2.44 MB/s) - ‘data/rdp_16s_v16.fa.gz’ saved [4602323/4602323]\n", | |
| "\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!sh code/download.sh" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 23, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "total 56M\r\n", | |
| "drwxr-xr-x 2 amsa amsa 4.0K Aug 14 2014 MiSeq_SOP\r\n", | |
| "-rw-rw-r-- 1 amsa amsa 36M Aug 14 2014 MiSeqSOPData.zip\r\n", | |
| "-rw-rw-r-- 1 amsa amsa 20M Sep 10 01:16 rdp_16s_v16.fa\r\n", | |
| "-rw-rw-r-- 1 amsa amsa 206 Jan 11 10:22 README\r\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!rm -r data/__MACOSX\n", | |
| "!ls -lh data" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "# Sequence preprocessing\n", | |
| "\n", | |
| "### Step 01: Merging pairs\n", | |
| "\n", | |
| "http://drive5.com/usearch/manual/cmd_fastq_mergepairs.html\n", | |
| "\n", | |
| "The fastq_mergepairs command merges (assembles) paired-end reads to create consensus sequences and, optionally, consensus quality scores.\n", | |
| "\n", | |
| "If given the forward reads eg. `F3D*_R1_*.fastq` the script automatically finds the R2 reads and matches the samples\n", | |
| "\n", | |
| "---\n", | |
| "\n", | |
| "After the merge examine the [merge report](http://drive5.com/usearch/manual/merge_report.html), which is part of the output.\n", | |
| "\n", | |
| "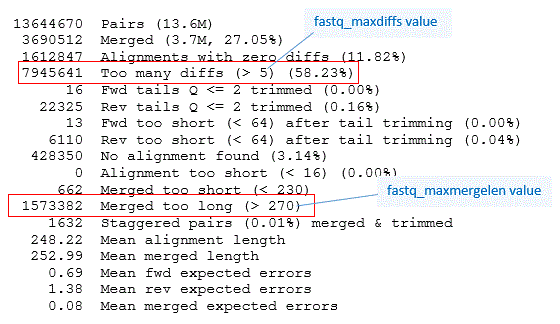\n", | |
| "\n", | |
| "With long overlaps (here shown by the mean alignment length of 248). Misalignments are therefore very unlikely, and it would be reasonable to increase the -fastq_maxdiffs and -fastq_maxdiffspct values to increase the number of merged pairs. Quality filtering will take care of discarding reads where many mismatches induce a large number of expected errors. This doesn't necessarily happen -- e.g., if low quality base calls in R2 are mismatches against high-quality base calls in R1 then the merged Q scores can still be high.\n", | |
| "\n", | |
| "**Filtering**\n", | |
| "\n", | |
| "`-fastq_maxdiffs` Maximum number of mismatches in the alignment. Default 5. Consider increasing if you have long overlaps.\n", | |
| "`-fastq_maxdiffpct` Maximum number of mismatches as an integer percentage. Default 5. Consider increasing if you have long overlaps.\n", | |
| "\n", | |
| "Deafult params work well for merging 2 x 300 reads with short overlap shuch as the V3-V4 product.\n", | |
| "\n", | |
| "**Merged read labels**\n", | |
| "\n", | |
| " `-relabel` Prefix string from sample file for output labels, then the read number 1, 2, 3... is appended after the prefix.\n", | |
| " \n", | |
| "`F3D9_S197_L001_R1_001.fastq` becomes\n", | |
| "\n", | |
| " @F3D9.1\n", | |
| " @F3D9.2\n", | |
| " @F3D9.3 ...\n" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 51, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [], | |
| "source": [ | |
| "!rm -r uparse_out\n", | |
| "!mkdir uparse_out" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 52, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "usearch v9.2.64_i86linux32, 4.0Gb RAM (8.1Gb total), 4 cores\n", | |
| "(C) Copyright 2013-16 Robert C. Edgar, all rights reserved.\n", | |
| "http://drive5.com/usearch\n", | |
| "\n", | |
| "License: amsa@eaaa.dk\n", | |
| "\n", | |
| "\n", | |
| "Merging\n", | |
| " Fwd data/MiSeq_SOP/F3D0_S188_L001_R1_001.fastq\n", | |
| " Rev data/MiSeq_SOP/F3D0_S188_L001_R2_001.fastq\n", | |
| " Relabel reads as F3D0.#\n", | |
| "\n", | |
| "00:00 100Mb 100.0% 84.8% merged\n", | |
| " \n", | |
| "Merging\n", | |
| " Fwd data/MiSeq_SOP/F3D141_S207_L001_R1_001.fastq\n", | |
| " Rev data/MiSeq_SOP/F3D141_S207_L001_R2_001.fastq\n", | |
| " Relabel reads as F3D141.#\n", | |
| "\n", | |
| "00:00 100Mb 0.1% 0% merged00:00 100Mb 100.0% 83.9% merged\n", | |
| " \n", | |
| "Merging\n", | |
| " Fwd data/MiSeq_SOP/F3D142_S208_L001_R1_001.fastq\n", | |
| " Rev data/MiSeq_SOP/F3D142_S208_L001_R2_001.fastq\n", | |
| " Relabel reads as F3D142.#\n", | |
| "\n", | |
| "00:01 100Mb 0.2% 83.9% merged00:01 100Mb 100.0% 83.3% merged\n", | |
| " \n", | |
| "Merging\n", | |
| " Fwd data/MiSeq_SOP/F3D143_S209_L001_R1_001.fastq\n", | |
| " Rev data/MiSeq_SOP/F3D143_S209_L001_R2_001.fastq\n", | |
| " Relabel reads as F3D143.#\n", | |
| "\n", | |
| "00:01 100Mb 0.1% 0% merged00:01 100Mb 100.0% 83.1% merged\n", | |
| " \n", | |
| "Merging\n", | |
| " Fwd data/MiSeq_SOP/F3D144_S210_L001_R1_001.fastq\n", | |
| " Rev data/MiSeq_SOP/F3D144_S210_L001_R2_001.fastq\n", | |
| " Relabel reads as F3D144.#\n", | |
| "\n", | |
| "00:01 100Mb 0.1% 0% merged00:01 100Mb 100.0% 81.9% merged\n", | |
| " \n", | |
| "Merging\n", | |
| " Fwd data/MiSeq_SOP/F3D145_S211_L001_R1_001.fastq\n", | |
| " Rev data/MiSeq_SOP/F3D145_S211_L001_R2_001.fastq\n", | |
| " Relabel reads as F3D145.#\n", | |
| "\n", | |
| "00:01 100Mb 0.1% 0% merged00:01 100Mb 100.0% 80.8% merged\n", | |
| " \n", | |
| "Merging\n", | |
| " Fwd data/MiSeq_SOP/F3D146_S212_L001_R1_001.fastq\n", | |
| " Rev data/MiSeq_SOP/F3D146_S212_L001_R2_001.fastq\n", | |
| " Relabel reads as F3D146.#\n", | |
| "\n", | |
| "00:01 100Mb 0.1% 0% merged00:01 100Mb 100.0% 81.2% merged\n", | |
| " \n", | |
| "Merging\n", | |
| " Fwd data/MiSeq_SOP/F3D147_S213_L001_R1_001.fastq\n", | |
| " Rev data/MiSeq_SOP/F3D147_S213_L001_R2_001.fastq\n", | |
| " Relabel reads as F3D147.#\n", | |
| "\n", | |
| "00:01 100Mb 0.1% 0% merged00:01 100Mb 100.0% 79.4% merged\n", | |
| " \n", | |
| "Merging\n", | |
| " Fwd data/MiSeq_SOP/F3D148_S214_L001_R1_001.fastq\n", | |
| " Rev data/MiSeq_SOP/F3D148_S214_L001_R2_001.fastq\n", | |
| " Relabel reads as F3D148.#\n", | |
| "\n", | |
| "00:01 100Mb 0.1% 0% merged00:01 100Mb 100.0% 79.6% merged\n", | |
| " \n", | |
| "Merging\n", | |
| " Fwd data/MiSeq_SOP/F3D149_S215_L001_R1_001.fastq\n", | |
| " Rev data/MiSeq_SOP/F3D149_S215_L001_R2_001.fastq\n", | |
| " Relabel reads as F3D149.#\n", | |
| "\n", | |
| "00:02 100Mb 0.2% 79.6% merged00:02 100Mb 100.0% 80.1% merged\n", | |
| " \n", | |
| "Merging\n", | |
| " Fwd data/MiSeq_SOP/F3D150_S216_L001_R1_001.fastq\n", | |
| " Rev data/MiSeq_SOP/F3D150_S216_L001_R2_001.fastq\n", | |
| " Relabel reads as F3D150.#\n", | |
| "\n", | |
| "00:02 100Mb 0.1% 0% merged00:02 100Mb 100.0% 80.1% merged\n", | |
| " \n", | |
| "Merging\n", | |
| " Fwd data/MiSeq_SOP/F3D1_S189_L001_R1_001.fastq\n", | |
| " Rev data/MiSeq_SOP/F3D1_S189_L001_R2_001.fastq\n", | |
| " Relabel reads as F3D1.#\n", | |
| "\n", | |
| "00:02 100Mb 0.1% 0% merged00:02 100Mb 100.0% 80.5% merged\n", | |
| " \n", | |
| "Merging\n", | |
| " Fwd data/MiSeq_SOP/F3D2_S190_L001_R1_001.fastq\n", | |
| " Rev data/MiSeq_SOP/F3D2_S190_L001_R2_001.fastq\n", | |
| " Relabel reads as F3D2.#\n", | |
| "\n", | |
| "00:02 100Mb 0.1% 0% merged00:02 101Mb 100.0% 81.3% merged\n", | |
| " \n", | |
| "Merging\n", | |
| " Fwd data/MiSeq_SOP/F3D3_S191_L001_R1_001.fastq\n", | |
| " Rev data/MiSeq_SOP/F3D3_S191_L001_R2_001.fastq\n", | |
| " Relabel reads as F3D3.#\n", | |
| "\n", | |
| "00:02 101Mb 0.1% 0% merged00:03 101Mb 48.1% 81.3% merged00:03 101Mb 100.0% 81.3% merged\n", | |
| " \n", | |
| "Merging\n", | |
| " Fwd data/MiSeq_SOP/F3D5_S193_L001_R1_001.fastq\n", | |
| " Rev data/MiSeq_SOP/F3D5_S193_L001_R2_001.fastq\n", | |
| " Relabel reads as F3D5.#\n", | |
| "\n", | |
| "00:03 101Mb 0.1% 0% merged00:03 104Mb 100.0% 81.3% merged\n", | |
| " \n", | |
| "Merging\n", | |
| " Fwd data/MiSeq_SOP/F3D6_S194_L001_R1_001.fastq\n", | |
| " Rev data/MiSeq_SOP/F3D6_S194_L001_R2_001.fastq\n", | |
| " Relabel reads as F3D6.#\n", | |
| "\n", | |
| "00:03 104Mb 0.1% 0% merged00:03 104Mb 100.0% 81.6% merged\n", | |
| " \n", | |
| "Merging\n", | |
| " Fwd data/MiSeq_SOP/F3D7_S195_L001_R1_001.fastq\n", | |
| " Rev data/MiSeq_SOP/F3D7_S195_L001_R2_001.fastq\n", | |
| " Relabel reads as F3D7.#\n", | |
| "\n", | |
| "00:03 104Mb 0.1% 0% merged00:03 104Mb 100.0% 81.6% merged\n", | |
| " \n", | |
| "Merging\n", | |
| " Fwd data/MiSeq_SOP/F3D8_S196_L001_R1_001.fastq\n", | |
| " Rev data/MiSeq_SOP/F3D8_S196_L001_R2_001.fastq\n", | |
| " Relabel reads as F3D8.#\n", | |
| "\n", | |
| "00:03 104Mb 0.1% 0% merged00:03 104Mb 100.0% 81.8% merged\n", | |
| " \n", | |
| "Merging\n", | |
| " Fwd data/MiSeq_SOP/F3D9_S197_L001_R1_001.fastq\n", | |
| " Rev data/MiSeq_SOP/F3D9_S197_L001_R2_001.fastq\n", | |
| " Relabel reads as F3D9.#\n", | |
| "\n", | |
| "00:03 104Mb 0.1% 0% merged00:03 104Mb 100.0% 82.1% merged\n", | |
| "\n", | |
| "Totals:\n", | |
| " 147581 Pairs (147.6k)\n", | |
| " 121176 Merged (121.2k, 82.11%)\n", | |
| " 40086 Alignments with zero diffs (27.16%)\n", | |
| " 26292 Too many diffs (> 10) (17.82%)\n", | |
| " 0 Fwd tails Q <= 2 trimmed (0.00%)\n", | |
| " 148 Rev tails Q <= 2 trimmed (0.10%)\n", | |
| " 0 Fwd too short (< 64) after tail trimming (0.00%)\n", | |
| " 5 Rev too short (< 64) after tail trimming (0.00%)\n", | |
| " 108 No alignment found (0.07%)\n", | |
| " 0 Alignment too short (< 16) (0.00%)\n", | |
| " 22 Staggered pairs (0.01%) merged & trimmed\n", | |
| " 249.23 Mean alignment length\n", | |
| " 252.47 Mean merged length\n", | |
| " 0.39 Mean fwd expected errors\n", | |
| " 1.14 Mean rev expected errors\n", | |
| " 0.08 Mean merged expected errors\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!usearch -fastq_mergepairs data/MiSeq_SOP/F3D*_R1_*.fastq -fastqout 'uparse_out/01_merged.fq' -relabel @ -log 'uparse_out/01_merge.log' -fastq_maxdiffs 10 -fastq_maxdiffpct 10" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 53, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| " Relabel reads as F3D9.#\r\n", | |
| "\r\n", | |
| "00:03 104Mb 100.0% 82.1% merged\r\n", | |
| "\r\n", | |
| "Totals:\r\n", | |
| " 147581 Pairs (147.6k)\r\n", | |
| " 121176 Merged (121.2k, 82.11%)\r\n", | |
| " 40086 Alignments with zero diffs (27.16%)\r\n", | |
| " 26292 Too many diffs (> 10) (17.82%)\r\n", | |
| " 0 Fwd tails Q <= 2 trimmed (0.00%)\r\n", | |
| " 148 Rev tails Q <= 2 trimmed (0.10%)\r\n", | |
| " 0 Fwd too short (< 64) after tail trimming (0.00%)\r\n", | |
| " 5 Rev too short (< 64) after tail trimming (0.00%)\r\n", | |
| " 108 No alignment found (0.07%)\r\n", | |
| " 0 Alignment too short (< 16) (0.00%)\r\n", | |
| " 22 Staggered pairs (0.01%) merged & trimmed\r\n", | |
| " 249.23 Mean alignment length\r\n", | |
| " 252.47 Mean merged length\r\n", | |
| " 0.39 Mean fwd expected errors\r\n", | |
| " 1.14 Mean rev expected errors\r\n", | |
| " 0.08 Mean merged expected errors\r\n", | |
| "\r\n", | |
| "Finished Wed Jan 11 12:02:31 2017\r\n", | |
| "Elapsed time 00:03\r\n", | |
| "Max memory 104Mb\r\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!tail -n 25 'uparse_out/01_merge.log'" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 54, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "@F3D0.1\r\n", | |
| "TACGGAGGATGCGAGCGTTATCCGGATTTATTGGGTTTAAAGGGTGCGTAGGCGGCCTGCCAAGTCAGCGGTAAAATTGCGGGGCTCAACCCCGTACAGCCGTTGAAACTGCCGGGCTCGAGTGGGCGAGAAGTATGCGGAATGCGTGGTGTAGCGGTGAAATGCATAGATATCACGCAGAACCCCGATTGCGAAGGCAGCATACCGGCGCCCTACTGACGCTGAGGCACGAAAGTGCGGGGATCAAACAGG\r\n", | |
| "+\r\n", | |
| "35JJJJJJJJJJJJJJ/JJJ5JJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJB\r\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!head -n 4 'uparse_out/01_merged.fq'" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "!! Need a quality check here" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "### Step 02: filter fastq\n", | |
| "http://drive5.com/usearch/manual/upp_readprep.html\n", | |
| "\n", | |
| "**Quality filtering**\n", | |
| "\n", | |
| "Quality filtering of FASTQ reads is done by the `fastq_filter` command. This enables length trimming and quality filtering to be done in a single step. If you have overlapping paired reads, then you probably don't need to trim the length.\n", | |
| "\n", | |
| "`-fastq_maxee`: set an expected error threshold (1.0 is recommended as a default). \n", | |
| "\n", | |
| "Don't worry if this filter appears to be stringent; you will probably find that most of the discarded reads map to OTUs after clustering.\n", | |
| "\n", | |
| "**Length trimming**\n", | |
| "\n", | |
| "`-fastq_trunclen`: If you have variable-length reads, such as 454, then you should trim them to a fixed length. Not a problem for Illumina unless the reads are truncated on quality score (above rejects reads based on average quality) and this is usually not a problem with overlapping reads." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 55, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "usearch v9.2.64_i86linux32, 4.0Gb RAM (8.1Gb total), 4 cores\n", | |
| "(C) Copyright 2013-16 Robert C. Edgar, all rights reserved.\n", | |
| "http://drive5.com/usearch\n", | |
| "\n", | |
| "License: amsa@eaaa.dk\n", | |
| "\n", | |
| "00:02 32Mb 100.0% Filtering, 98.9% passed00:02 66Mb 98.9% Filtering, 98.9% passed\n", | |
| " 121176 Reads (121.2k) \n", | |
| " 1370 Discarded reads with expected errs > 1.00\n", | |
| " 119806 Filtered reads (119.8k, 98.9%)\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!usearch -fastq_filter 'uparse_out/01_merged.fq' -fastq_maxee 1.0 -fastaout 'uparse_out/02_filtered.fa' -relabel Filt -log 'uparse_out/02_filter.log'" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "These data are high quality, paired end reads that have been merged so only 1.1% failed. " | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 56, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| ">Filt1\r\n", | |
| "TACGGAGGATGCGAGCGTTATCCGGATTTATTGGGTTTAAAGGGTGCGTAGGCGGCCTGCCAAGTCAGCGGTAAAATTGC\r\n", | |
| "GGGGCTCAACCCCGTACAGCCGTTGAAACTGCCGGGCTCGAGTGGGCGAGAAGTATGCGGAATGCGTGGTGTAGCGGTGA\r\n", | |
| "AATGCATAGATATCACGCAGAACCCCGATTGCGAAGGCAGCATACCGGCGCCCTACTGACGCTGAGGCACGAAAGTGCGG\r\n", | |
| "GGATCAAACAGG\r\n", | |
| ">Filt2\r\n", | |
| "TACGGAGGATGCGAGCGTTATCCGGATTTATTGGGTTTAAAGGGTGCGCAGGCGGAAGATCAAGTCAGCGGTAAAATTGA\r\n", | |
| "GAGGCTCAACCTCTTCGAGCCGTTGAAACTGGTTTTCTTGAGTGAGCGAGAAGTATGCGGAATGCGTGGTGTAGCGGTGA\r\n", | |
| "AATGCATAGATATCACGCAGAACTCCGATTGCGAAGGCAGCATACCGGCGCTCAACTGACGCTCATGCACGAAAGTGTGG\r\n", | |
| "GTATCGAACAGG\r\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!head 'uparse_out/02_filtered.fa'" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "### Step 03: Dereplication\n", | |
| "\n", | |
| "The input sequences to the first `cluster_otus` (that generates the zOTUs) must be a set of unique sequences sorted in order of decreasing abundance with size annotations in the labels. The `fastx_uniques` command can do this. The input to `fastx_uniques` should be the reads after any quality filtering or length trimming.\n", | |
| "\n", | |
| "`-relabel Uniq` so that the unique sequences are labeled `Uniq1, Uniq2` and so on. \n", | |
| "\n", | |
| "**Samples should be pooled before dereplication**\n", | |
| "\n", | |
| "I recommend pooling samples, i.e. concatenating reads for all samples that were sequenced in the same run. This is important for getting the best detection of chimeras and cross-talk, and for getting the best sensitivity to low-abundance sequences that could be lost if individual samples or subsets of samples are clustered separately. See pooling samples for discussion." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 57, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "usearch v9.2.64_i86linux32, 4.0Gb RAM (8.1Gb total), 4 cores\n", | |
| "(C) Copyright 2013-16 Robert C. Edgar, all rights reserved.\n", | |
| "http://drive5.com/usearch\n", | |
| "\n", | |
| "License: amsa@eaaa.dk\n", | |
| "\n", | |
| "00:00 73Mb 100.0% Reading uparse_out/02_filtered.fa\n", | |
| "00:01 78Mb 100.0% DF 00:01 78Mb 68.2% DF\n", | |
| "00:01 80Mb 119806 seqs, 15780 uniques, 12315 singletons (78.0%)\n", | |
| "00:01 80Mb Min size 1, median 1, max 9798, avg 7.59\n", | |
| "00:01 72Mb 0.0% Writing uparse_out/03_uniques.fa00:01 72Mb 100.0% Writing uparse_out/03_uniques.fa\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!usearch -fastx_uniques 'uparse_out/02_filtered.fa' -sizeout -relabel Uniq -fastaout 'uparse_out/03_uniques.fa' -log 'uparse_out/03_uniques.log'" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 58, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| ">Uniq1;size=9798;\r\n", | |
| "TACGGAGGATGCGAGCGTTATCCGGATTTATTGGGTTTAAAGGGTGCGCAGGCGGAAGATCAAGTCAGCGGTAAAATTGA\r\n", | |
| "GAGGCTCAACCTCTTCGAGCCGTTGAAACTGGTTTTCTTGAGTGAGCGAGAAGTATGCGGAATGCGTGGTGTAGCGGTGA\r\n", | |
| "AATGCATAGATATCACGCAGAACTCCGATTGCGAAGGCAGCATACCGGCGCTCAACTGACGCTCATGCACGAAAGTGTGG\r\n", | |
| "GTATCGAACAGG\r\n", | |
| ">Uniq2;size=7743;\r\n", | |
| "TACGGAGGATGCGAGCGTTATCCGGATTTATTGGGTTTAAAGGGTGCGCAGGCGGACTCTCAAGTCAGCGGTCAAATCGC\r\n", | |
| "GGGGCTCAACCCCGTTCCGCCGTTGAAACTGGGAGCCTTGAGTGCGCGAGAAGTAGGCGGAATGCGTGGTGTAGCGGTGA\r\n", | |
| "AATGCATAGATATCACGCAGAACTCCGATTGCGAAGGCAGCCTACCGGCGCGCAACTGACGCTCATGCACGAAAGCGTGG\r\n", | |
| "GTATCGAACAGG\r\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!head 'uparse_out/03_uniques.fa'" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "# UPARSE\n", | |
| "\n", | |
| "\n", | |
| "### Step 04: : Identify centroids and chimera filter: UCHIME\n", | |
| "\n", | |
| "The purpose of this step is to identify the zOTUs in the population. The high-abundance, good quality sequences that we assume are the centroids of the actual population OTUs. In a UPARSE pipeline, clustering and chimera filtering is performed by the `cluster_otus` command. \n", | |
| "\n", | |
| "The chimera filter in cluster_otus is very effective, so I generally don't do any additional filtering. It may be possible to find some additional chimeras by running uchime2_ref on the OTU sequences generated by `cluster_otus`.\n", | |
| "\n", | |
| "A typical command line is:\n", | |
| "\n", | |
| " usearch -cluster_otus derep.fa -otus otus.fa -relabel OTU -minsize 2 -uparseout out.up \n", | |
| "\n", | |
| "`-otu_radius_pct` Default is 3.0, corresponding to a minimum identity of 97%.\n", | |
| "\n", | |
| "`-otus` specifies a FASTA filename for the OTU representative sequences. By default, labels are taken from the input file, with size annotations stripped. \n", | |
| "\n", | |
| "`-relabel` option specifies a string that is used to re-label OTUs. If -relabel xxx is specified, then the labels are xxx followed by 1, 2 ... up to the number of OTUs. OTU identifiers in the labels is required for making an OTU table using usearch_global\n", | |
| "\n", | |
| "`‑minsize 2` option discards singleton unique sequences, most of which will contain sequencer error. See discarding singletons for discussion. Most singletons will be assigned to OTUs when the OTU table is constructed, so the data is not lost.\n", | |
| "\n", | |
| "\n", | |
| "Optional:\n", | |
| "\n", | |
| "`‑mode high_confidence` can be used to reduce the risk of false positive chimeras.\n", | |
| "\n", | |
| "`-uparseout` specifies a tabbed text output file documenting how the input sequences were classified." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 59, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "usearch v9.2.64_i86linux32, 4.0Gb RAM (8.1Gb total), 4 cores\n", | |
| "(C) Copyright 2013-16 Robert C. Edgar, all rights reserved.\n", | |
| "http://drive5.com/usearch\n", | |
| "\n", | |
| "License: amsa@eaaa.dk\n", | |
| "\n", | |
| "00:01 47Mb 6.5% 183 OTUs, 59 chimeras00:01 48Mb 100.0% 207 OTUs, 165 chimeras\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!usearch -cluster_otus 'uparse_out/03_uniques.fa' -minsize 2 -otus 'uparse_out/04_otus.fa' -relabel Otu -log 'uparse_out/04_cluster_otus.log'" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "### Classify OTUs: SINTAX\n", | |
| "\n", | |
| "Classify the centroids (repseqs)." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 60, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "usearch v9.2.64_i86linux32, 4.0Gb RAM (8.1Gb total), 4 cores\n", | |
| "(C) Copyright 2013-16 Robert C. Edgar, all rights reserved.\n", | |
| "http://drive5.com/usearch\n", | |
| "\n", | |
| "License: amsa@eaaa.dk\n", | |
| "\n", | |
| "00:00 61Mb 100.0% Reading data/rdp_16s_v16.fa\n", | |
| "00:00 27Mb 0.1% Masking (fastnucleo) 00:01 27Mb 76.8% Masking (fastnucleo)00:01 27Mb 100.0% Masking (fastnucleo)\n", | |
| "00:02 28Mb 100.0% Word stats 00:02 28Mb 98.4% Word stats\n", | |
| "00:02 28Mb 100.0% Alloc rows\n", | |
| "00:02 99Mb 0.1% Build index00:03 99Mb 64.8% Build index00:03 99Mb 100.0% Build index\n", | |
| "00:03 99Mb 0.0% Initialize taxonomy data00:03 100Mb 100.0% Initialize taxonomy data\n", | |
| "00:03 100Mb 100.0% Building name table \n", | |
| "00:03 100Mb 3172 names, tax levels min 3, avg 5.9, max 6\n", | |
| "00:03 133Mb 0.1% Processing00:04 163Mb 20.3% Processing00:05 163Mb 75.3% Processing00:05 163Mb 100.0% Processing\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!usearch -sintax 'uparse_out/04_otus.fa' -db data/rdp_16s_v16.fa -strand plus -tabbedout 'uparse_out/04_otus_sintax.txt' -log 'uparse_out/04_sintax.log'" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "### Step 05: Populate OTU tables with clustering at 97% identity\n", | |
| "\n", | |
| "http://drive5.com/usearch/manual/mapreadstootus.html\n", | |
| "\n", | |
| "Now that the high quality centroids have been identified the surrounding _member_ (point error) reads can then be clustered with each centroid." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 61, | |
| "metadata": { | |
| "collapsed": false, | |
| "scrolled": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "usearch v9.2.64_i86linux32, 4.0Gb RAM (8.1Gb total), 4 cores\n", | |
| "(C) Copyright 2013-16 Robert C. Edgar, all rights reserved.\n", | |
| "http://drive5.com/usearch\n", | |
| "\n", | |
| "License: amsa@eaaa.dk\n", | |
| "\n", | |
| "00:01 56Mb 76.7% Reading data/rdp_16s_v16.fa00:01 61Mb 100.0% Reading data/rdp_16s_v16.fa\n", | |
| "00:01 28Mb 0.1% Masking (fastnucleo) 00:01 28Mb 100.0% Masking (fastnucleo)\n", | |
| "00:01 28Mb 0.1% Word stats 00:02 28Mb 54.6% Word stats00:02 28Mb 100.0% Word stats\n", | |
| "00:02 28Mb 100.0% Alloc rows\n", | |
| "00:02 99Mb 0.1% Build index00:03 99Mb 34.9% Build index00:04 99Mb 95.3% Build index00:04 99Mb 100.0% Build index\n", | |
| "00:04 132Mb 0.1% Searching 04_otus.fa, 0.0% matched00:04 166Mb 100.0% Searching 04_otus.fa, 14.5% matched\n", | |
| "usearch v9.2.64_i86linux32, 4.0Gb RAM (8.1Gb total), 4 cores\n", | |
| "(C) Copyright 2013-16 Robert C. Edgar, all rights reserved.\n", | |
| "http://drive5.com/usearch\n", | |
| "\n", | |
| "License: amsa@eaaa.dk\n", | |
| "\n", | |
| "00:00 40Mb 100.0% Reading uparse_out/04_otus.fa\n", | |
| "00:00 6.5Mb 100.0% Masking (fastnucleo) \n", | |
| "00:00 7.5Mb 100.0% Word stats \n", | |
| "00:00 7.5Mb 100.0% Alloc rows\n", | |
| "00:00 7.7Mb 100.0% Build index\n", | |
| "00:00 41Mb 0.1% Searching 01_merged.fq, 0.0% matched00:01 74Mb 6.7% Searching 01_merged.fq, 96.9% matched00:02 74Mb 41.6% Searching 01_merged.fq, 96.0% matched00:03 74Mb 92.2% Searching 01_merged.fq, 96.4% matched00:03 74Mb 100.0% Searching 01_merged.fq, 96.5% matched\n", | |
| "116831 / 121176 mapped to OTUs (96.4%) \n", | |
| "00:03 74Mb Writing uparse_out/05_otutab.txt\n", | |
| "00:03 74Mb Writing uparse_out/05_otutab.txt ...done.\n", | |
| "00:03 74Mb Writing uparse_out/05_otutab.json\n", | |
| "00:03 74Mb Writing uparse_out/05_otutab.json ...done.\n", | |
| "00:03 74Mb Writing uparse_out/05_otutab.mothur\n", | |
| "00:03 74Mb Writing uparse_out/05_otutab.mothur ...done.\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!usearch -usearch_global 'uparse_out/04_otus.fa' -db 'data/rdp_16s_v16.fa' -strand plus -id 0.97 -alnout 'uparse_out/04_otus_ref.aln' -userout 'uparse_out/04_otus_ref.user' -userfields query+target+id\n", | |
| "\n", | |
| "!usearch -usearch_global 'uparse_out/01_merged.fq' -db 'uparse_out/04_otus.fa' -strand plus -id 0.97 -log 'uparse_out/05_make_otutab.log' -otutabout 'uparse_out/05_otutab.txt' -biomout 'uparse_out/05_otutab.json' -mothur_shared_out 'uparse_out/05_otutab.mothur'" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "### Step 06: Remove sequencing errors: UNOISE2\n", | |
| "\n", | |
| "http://www.drive5.com/usearch/manual/cmd_unoise2.html\n", | |
| "\n", | |
| "Edgar, R.C. (2016), UNOISE2: Improved error-correction for Illumina 16S and ITS amplicon reads.http://dx.doi.org/10.1101/081257\n", | |
| "\n", | |
| "#### UNOISE algorithm\n", | |
| "\n", | |
| "**Skew**\n", | |
| "\n", | |
| "Skew is the relative abundance of member sequence $M$ (abundance $a_M$) and a centroid ($C$, abundance $a_C$).\n", | |
| "\n", | |
| "$skew(M, C) = a_M / a_C$\n", | |
| "\n", | |
| "**distance**\n", | |
| "\n", | |
| "$d$ is the Levenshtein distance (n diffs inc. substitutions and gaps) between $C$ & $M$\n", | |
| "\n", | |
| "\n", | |
| "**Error model**\n", | |
| "\n", | |
| "A member $M$ is is probably an erroneous sequence derived from $C$ when it has:\n", | |
| "\n", | |
| "1. It is in low enough abundance compared to $C$ => small enough $skew$ with respect to $M$ (higher $skew$ more likely an error)\n", | |
| "2. small enough $d$ (more differences => higher $d$ => more likely an error)\n", | |
| "\n", | |
| "This is formalised in a distribution model where $\\beta$ is the maximum skew allowed for a member ($M$) with $d$ differences:\n", | |
| "\n", | |
| "$\\beta(d) = 1 / 2^{\\alpha d + 1}$\n", | |
| "\n", | |
| "\n", | |
| "where: $\\alpha$ = 2 (default). \n", | |
| "\n", | |
| "$\\beta(1) = 1/8, \\beta(2) = 1/32, \\beta(3) = 1/128 ... $\n", | |
| "\n", | |
| "So if $skew(M, C) >= \\beta(d)$ then the $M$ is an error, and $\\beta$ increases exponentially with $d$.\n", | |
| "\n", | |
| "Higher $\\alpha$ means you get more \"real\" sequences but increases in the number of bad sequences that are assumed to be good. \n", | |
| "\n", | |
| "\n", | |
| "Where $\\alpha$ = 3. \n", | |
| "\n", | |
| "$\\beta(1) = 1/16, \\beta(2) = 1/128, \\beta(3) = 1/1024 ... $ \n", | |
| "\n", | |
| "so sequences with $d = 1$ and $skew(M, C)$ between 1/8 and 1/16 are assumed errors when $\\alpha$ = 2, but accepted as \"real\" when $\\alpha$ = 3.\n", | |
| "\n", | |
| "---\n", | |
| "\n", | |
| "The input to UNOISE2 is the good quality, unique reads from preprocessing with size=nnn; abundance annotations. \n", | |
| "\n", | |
| "It uses a one-pass clustering that does not use quality scores (Q).\n", | |
| "\n", | |
| "It has two parameters with preset values:\n", | |
| "\n", | |
| "$\\alpha$ = 2 (default) is the sensitivity to difference in abundance with a centroid\n", | |
| "$\\gamma$ = 4 (default) is a threshold that sets the the minimum number of reads with a given unique sequence. A smaller $\\gamma$ will increase the number of spurious zOTUs. \n", | |
| "\n", | |
| "It is also therefore better to pool samples sa that there is more data and if they fail it is due to the $\\alpha$ nit the $\\gamma$.\n", | |
| "\n", | |
| "\n", | |
| "Errors are corrected as follows:\n", | |
| " - Reads with sequencing error are identified and corrected.\n", | |
| " - Chimeras are removed.\n", | |
| " - PhiX sequences are removed.\n", | |
| " \n", | |
| "`-fastaout` denoised sequences (\"Corrected biological sequences\"). \n", | |
| "\n", | |
| "Labels are formatted as Otunnn;Uniqlabel; where nnn is 1, 2, 3... and Uniqlabel is the label from the input file (truncated at the first semi-colon, to strip any annotations).\n", | |
| "\n", | |
| "\n", | |
| "`-minampsize` option specifies the minimum abundance (size= annotation) for an error-corrected amplicon. Default is 4.\n" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 62, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "usearch v9.2.64_i86linux32, 4.0Gb RAM (8.1Gb total), 4 cores\n", | |
| "(C) Copyright 2013-16 Robert C. Edgar, all rights reserved.\n", | |
| "http://drive5.com/usearch\n", | |
| "\n", | |
| "License: amsa@eaaa.dk\n", | |
| "\n", | |
| "00:00 208Mb 100.0% Word stats\n", | |
| "00:00 208Mb 100.0% Alloc rows\n", | |
| "00:00 208Mb 100.0% Build index\n", | |
| "00:01 175Mb 0.1% Reading uparse_out/03_uniques.fa00:01 182Mb 100.0% Reading uparse_out/03_uniques.fa\n", | |
| "00:01 150Mb 0.1% 0 amplicons, 0 bad (size >= 9798)00:01 159Mb 100.0% 607 amplicons, 6259 bad (size >= 4)\n", | |
| "00:01 162Mb 0.2% 1 good, 0 chimeras 00:02 166Mb 64.1% 254 good, 136 chimeras00:03 166Mb 89.0% 296 good, 245 chimeras00:03 166Mb 100.0% 303 good, 305 chimeras\n", | |
| "00:03 166Mb 100.0% Writing amplicons \n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!usearch -unoise2 'uparse_out/03_uniques.fa' -fastaout 'uparse_out/06_denoised.fa' -log 'uparse_out/06_unoise.log' -minampsize 4" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 63, | |
| "metadata": { | |
| "collapsed": false, | |
| "scrolled": true | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "usearch v9.2.64_i86linux32, 4.0Gb RAM (8.1Gb total), 4 cores\n", | |
| "(C) Copyright 2013-16 Robert C. Edgar, all rights reserved.\n", | |
| "http://drive5.com/usearch\n", | |
| "\n", | |
| "License: amsa@eaaa.dk\n", | |
| "\n", | |
| "00:00 61Mb 100.0% Reading data/rdp_16s_v16.fa\n", | |
| "00:00 27Mb 0.1% Masking (fastnucleo) 00:01 27Mb 28.2% Masking (fastnucleo)00:01 27Mb 100.0% Masking (fastnucleo)\n", | |
| "00:01 28Mb 0.1% Word stats 00:02 28Mb 65.6% Word stats00:02 28Mb 100.0% Word stats\n", | |
| "00:02 28Mb 100.0% Alloc rows\n", | |
| "00:02 99Mb 0.1% Build index00:03 99Mb 34.7% Build index00:03 99Mb 100.0% Build index\n", | |
| "00:03 99Mb 0.0% Initialize taxonomy data00:03 100Mb 100.0% Initialize taxonomy data\n", | |
| "00:03 100Mb 100.0% Building name table \n", | |
| "00:03 100Mb 3172 names, tax levels min 3, avg 5.9, max 6\n", | |
| "00:03 133Mb 0.1% Processing00:04 164Mb 6.0% Processing00:05 164Mb 56.4% Processing00:05 164Mb 100.0% Processing\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!usearch -sintax 'uparse_out/06_denoised.fa' -db data/rdp_16s_v16.fa -strand plus -tabbedout 'uparse_out/06_otus_sintax.txt' -log 'uparse_out/06_sintax.log'" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 64, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| ">Otu1;uniq=Uniq1;size=9798;\r\n", | |
| "TACGGAGGATGCGAGCGTTATCCGGATTTATTGGGTTTAAAGGGTGCGCAGGCGGAAGATCAAGTCAGCGGTAAAATTGA\r\n", | |
| "GAGGCTCAACCTCTTCGAGCCGTTGAAACTGGTTTTCTTGAGTGAGCGAGAAGTATGCGGAATGCGTGGTGTAGCGGTGA\r\n", | |
| "AATGCATAGATATCACGCAGAACTCCGATTGCGAAGGCAGCATACCGGCGCTCAACTGACGCTCATGCACGAAAGTGTGG\r\n", | |
| "GTATCGAACAGG\r\n", | |
| ">Otu2;uniq=Uniq2;size=7743;\r\n", | |
| "TACGGAGGATGCGAGCGTTATCCGGATTTATTGGGTTTAAAGGGTGCGCAGGCGGACTCTCAAGTCAGCGGTCAAATCGC\r\n", | |
| "GGGGCTCAACCCCGTTCCGCCGTTGAAACTGGGAGCCTTGAGTGCGCGAGAAGTAGGCGGAATGCGTGGTGTAGCGGTGA\r\n", | |
| "AATGCATAGATATCACGCAGAACTCCGATTGCGAAGGCAGCCTACCGGCGCGCAACTGACGCTCATGCACGAAAGCGTGG\r\n", | |
| "GTATCGAACAGG\r\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!head uparse_out/06_denoised.fa" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "### Step 07: Populate OTU table\n", | |
| "\n", | |
| "Search for one (default) or a few high-identity hits to a database using the USEARCH algorithm. Alignments are global. \n", | |
| "\n", | |
| "To get more than one hit, increase `-maxaccepts` (see accept options).\n", | |
| "\n", | |
| "`‑id` specifies the identity threshold. \n", | |
| "\n", | |
| "If full-length, exact matches are required, then it is better to use search_exact than usearch_global with -id 1.0." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 65, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "usearch v9.2.64_i86linux32, 4.0Gb RAM (8.1Gb total), 4 cores\n", | |
| "(C) Copyright 2013-16 Robert C. Edgar, all rights reserved.\n", | |
| "http://drive5.com/usearch\n", | |
| "\n", | |
| "License: amsa@eaaa.dk\n", | |
| "\n", | |
| "00:00 40Mb 100.0% Reading uparse_out/06_denoised.fa\n", | |
| "00:00 6.7Mb 100.0% Masking (fastnucleo) \n", | |
| "00:00 7.5Mb 100.0% Word stats \n", | |
| "00:00 7.5Mb 100.0% Alloc rows\n", | |
| "00:00 7.8Mb 100.0% Build index\n", | |
| "00:01 74Mb 38.3% Searching 01_merged.fq, 97.4% matched00:02 74Mb 89.0% Searching 01_merged.fq, 97.8% matched00:02 74Mb 100.0% Searching 01_merged.fq, 97.8% matched\n", | |
| "118456 / 121176 mapped to OTUs (97.8%) \n", | |
| "00:02 74Mb Writing uparse_out/07_otutab_den.txt\n", | |
| "00:02 74Mb Writing uparse_out/07_otutab_den.txt ...done.\n", | |
| "00:02 74Mb Writing uparse_out/07_otutab_den.json\n", | |
| "00:02 74Mb Writing uparse_out/07_otutab_den.json ...done.\n", | |
| "00:02 74Mb Writing uparse_out/07_otutab_den.mothur\n", | |
| "00:02 74Mb Writing uparse_out/07_otutab_den.mothur ...done.\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!usearch -usearch_global 'uparse_out/01_merged.fq' -db 'uparse_out/06_denoised.fa' -strand plus -id 0.97 -log 'uparse_out/07_make_otutab_den.log' -otutabout 'uparse_out/07_otutab_den.txt' -biomout 'uparse_out/07_otutab_den.json' -mothur_shared_out 'uparse_out/07_otutab_den.mothur'" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "## second option: Make otudb and use usearch_exact (faster)\n", | |
| "\n", | |
| "### Step 08: UNOISE\n", | |
| "\n", | |
| "`-otudbout` option specifies a FASTA filename which contains all non-chimeric unique sequences annotated with the Otu identifier of the corrected sequence. This enables generating a OTU table with `search_exact` rather than `usearch_global`, which is much faster than `usearch_global`. \n", | |
| "\n", | |
| "Sequences in the FASTA file are annotated as `Otun;Ampm;uniq=u;diffs=d`; To generate an OTU table, this file is used as a database and reads with sample identifiers are used as the query set. \n", | |
| "\n", | |
| "The `-otutabout`, `-biomout` and -`mothur_shared_out` options of `search_exact` specify OTU table file names. An OTU table can also be generated using `usearch_global`.\n" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 66, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "usearch v9.2.64_i86linux32, 4.0Gb RAM (8.1Gb total), 4 cores\n", | |
| "(C) Copyright 2013-16 Robert C. Edgar, all rights reserved.\n", | |
| "http://drive5.com/usearch\n", | |
| "\n", | |
| "License: amsa@eaaa.dk\n", | |
| "\n", | |
| "00:00 208Mb 100.0% Word stats\n", | |
| "00:00 208Mb 100.0% Alloc rows\n", | |
| "00:00 208Mb 100.0% Build index\n", | |
| "00:00 175Mb 0.1% Reading uparse_out/03_uniques.fa00:00 182Mb 100.0% Reading uparse_out/03_uniques.fa\n", | |
| "00:00 150Mb 0.1% 0 amplicons, 0 bad (size >= 9798)00:00 159Mb 100.0% 607 amplicons, 6259 bad (size >= 4)\n", | |
| "00:00 162Mb 0.2% 1 good, 0 chimeras 00:01 166Mb 18.8% 103 good, 11 chimeras00:02 166Mb 70.4% 266 good, 162 chimeras00:03 166Mb 91.0% 298 good, 255 chimeras00:03 166Mb 100.0% 303 good, 305 chimeras\n", | |
| "00:03 166Mb 100.0% Writing OTU db \n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!usearch -unoise2 'uparse_out/03_uniques.fa' -otudbout 'uparse_out/08_denoised_db.fa' -log 'uparse_out/08_unoise_db.log' -minampsize 4" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 67, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| ">Otu1;amp=Amp1;uniq=152886412;diffs=0;\r\n", | |
| "TACGGAGGATGCGAGCGTTATCCGGATTTATTGGGTTTAAAGGGTGCGCAGGCGGAAGATCAAGTCAGCGGTAAAATTGA\r\n", | |
| "GAGGCTCAACCTCTTCGAGCCGTTGAAACTGGTTTTCTTGAGTGAGCGAGAAGTATGCGGAATGCGTGGTGTAGCGGTGA\r\n", | |
| "AATGCATAGATATCACGCAGAACTCCGATTGCGAAGGCAGCATACCGGCGCTCAACTGACGCTCATGCACGAAAGTGTGG\r\n", | |
| "GTATCGAACAGG\r\n", | |
| ">Otu2;amp=Amp2;uniq=152886412;diffs=0;\r\n", | |
| "TACGGAGGATGCGAGCGTTATCCGGATTTATTGGGTTTAAAGGGTGCGCAGGCGGACTCTCAAGTCAGCGGTCAAATCGC\r\n", | |
| "GGGGCTCAACCCCGTTCCGCCGTTGAAACTGGGAGCCTTGAGTGCGCGAGAAGTAGGCGGAATGCGTGGTGTAGCGGTGA\r\n", | |
| "AATGCATAGATATCACGCAGAACTCCGATTGCGAAGGCAGCCTACCGGCGCGCAACTGACGCTCATGCACGAAAGCGTGG\r\n", | |
| "GTATCGAACAGG\r\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!head 'uparse_out/08_denoised_db.fa'" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "### Classify: SINTAX" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 68, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "usearch v9.2.64_i86linux32, 4.0Gb RAM (8.1Gb total), 4 cores\n", | |
| "(C) Copyright 2013-16 Robert C. Edgar, all rights reserved.\n", | |
| "http://drive5.com/usearch\n", | |
| "\n", | |
| "License: amsa@eaaa.dk\n", | |
| "\n", | |
| "00:01 57Mb 82.3% Reading data/rdp_16s_v16.fa00:01 61Mb 100.0% Reading data/rdp_16s_v16.fa\n", | |
| "00:01 27Mb 0.1% Masking (fastnucleo) 00:01 27Mb 100.0% Masking (fastnucleo)\n", | |
| "00:01 28Mb 0.1% Word stats 00:02 28Mb 38.9% Word stats00:02 28Mb 100.0% Word stats\n", | |
| "00:02 28Mb 100.0% Alloc rows\n", | |
| "00:02 99Mb 0.1% Build index00:03 99Mb 6.8% Build index00:04 99Mb 71.1% Build index00:04 99Mb 100.0% Build index\n", | |
| "00:04 99Mb 0.0% Initialize taxonomy data00:04 100Mb 100.0% Initialize taxonomy data\n", | |
| "00:04 100Mb 100.0% Building name table \n", | |
| "00:04 100Mb 3172 names, tax levels min 3, avg 5.9, max 6\n", | |
| "00:04 133Mb 0.1% Processing00:05 163Mb 5.1% Processing00:06 163Mb 17.6% Processing00:07 163Mb 28.4% Processing00:08 163Mb 44.4% Processing00:09 163Mb 58.7% Processing00:10 163Mb 70.8% Processing00:11 163Mb 85.0% Processing00:12 163Mb 98.5% Processing00:12 163Mb 100.0% Processing\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!usearch -sintax 'uparse_out/08_denoised_db.fa' -db data/rdp_16s_v16.fa -strand plus -tabbedout 'uparse_out/08_otus_sintax.txt' -log 'uparse_out/08_sintax.log'" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "### Step 09: Populate OTU table (search_exact)\n", | |
| "\n", | |
| "`search_exact` command searches for exact, full-length matches to a database sequence. Same as `usearch_global` with -id 1.0, but the algorithm is faster, uses less memory, and is guaranteed to find all correct matches, i.e. there are no heuristics.\n", | |
| "\n", | |
| "\n", | |
| "`-notmatched` output file can be used to save query sequences that are not found in the database\n", | |
| "\n", | |
| "`-dbnotmatched` option can be used to save database sequences that are not present in the query set. This enables incremental updating of dereplicated sequence sets." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 69, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "usearch v9.2.64_i86linux32, 4.0Gb RAM (8.1Gb total), 4 cores\n", | |
| "(C) Copyright 2013-16 Robert C. Edgar, all rights reserved.\n", | |
| "http://drive5.com/usearch\n", | |
| "\n", | |
| "License: amsa@eaaa.dk\n", | |
| "\n", | |
| "00:00 40Mb 100.0% Reading uparse_out/08_denoised_db.fa\n", | |
| "00:00 6.8Mb 100.0% Masking (fastnucleo) \n", | |
| "00:01 69Mb 61.5% Processing 00:01 69Mb 100.0% Processing\n", | |
| "96236 / 121176 mapped to OTUs (79.4%)\n", | |
| "00:01 69Mb Writing uparse_out/09_otutab_den.txt\n", | |
| "00:01 69Mb Writing uparse_out/09_otutab_den.txt ...done.\n", | |
| "00:01 69Mb Writing uparse_out/09_otutab_den.json\n", | |
| "00:01 69Mb Writing uparse_out/09_otutab_den.json ...done.\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!usearch -search_exact 'uparse_out/01_merged.fq' -db 'uparse_out/08_denoised_db.fa' -strand plus -log 'uparse_out/09_make_otutab_dendb.log' -otutabout 'uparse_out/09_otutab_den.txt' -biomout 'uparse_out/09_otutab_den.json' " | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Saved 2 seconds! OK it is a small dataset" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": 70, | |
| "metadata": { | |
| "collapsed": false | |
| }, | |
| "outputs": [ | |
| { | |
| "name": "stdout", | |
| "output_type": "stream", | |
| "text": [ | |
| "ls: cannot access 'out': No such file or directory\r\n" | |
| ] | |
| } | |
| ], | |
| "source": [ | |
| "!ls -lh out" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": null, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "execution_count": null, | |
| "metadata": { | |
| "collapsed": true | |
| }, | |
| "outputs": [], | |
| "source": [] | |
| } | |
| ], | |
| "metadata": { | |
| "anaconda-cloud": {}, | |
| "kernelspec": { | |
| "display_name": "Python [default]", | |
| "language": "python", | |
| "name": "python3" | |
| }, | |
| "language_info": { | |
| "codemirror_mode": { | |
| "name": "ipython", | |
| "version": 3 | |
| }, | |
| "file_extension": ".py", | |
| "mimetype": "text/x-python", | |
| "name": "python", | |
| "nbconvert_exporter": "python", | |
| "pygments_lexer": "ipython3", | |
| "version": "3.5.2" | |
| } | |
| }, | |
| "nbformat": 4, | |
| "nbformat_minor": 2 | |
| } |
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
please can you help me to merge all my forward and reverse files....usearch -fastq_mergepairs R1.fastq -relabel @ -fastqout merged.fq is not working on my windows.
It says cannot open R1