Created
October 15, 2014 13:37
-
-
Save allenday/f9d512dc5aae1a6ee204 to your computer and use it in GitHub Desktop.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| { | |
| "metadata": { | |
| "name": "", | |
| "signature": "sha256:69ee4419084fa9384e9f67f3928f7bad7602644de1278e93ed44ca70930b09cb" | |
| }, | |
| "nbformat": 3, | |
| "nbformat_minor": 0, | |
| "worksheets": [ | |
| { | |
| "cells": [ | |
| { | |
| "cell_type": "heading", | |
| "level": 1, | |
| "metadata": {}, | |
| "source": [ | |
| "Lesson 02: Probabilistic Data Structures" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "[Probabilistic Data Structures](http://highlyscalable.wordpress.com/2012/05/01/probabilistic-structures-web-analytics-data-mining/) are a fascinating and relatively new area of algorithms... largely pioneered by just a few individuals \u2013 e.g., [Philippe Flagolet](http://algo.inria.fr/flajolet/). Lately these have been finding oh so many uses, particularly for analytics with large-scale data and streaming use cases." | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "When you have a spare hour or two, definitely check out this O'Reilly webcast, [Probabilistic Data Structures and Breaking Down Big Sequence Data](http://www.oreilly.com/pub/e/1784) by C. Titus Brown:" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": true, | |
| "input": [ | |
| "import IPython.display\n", | |
| "import urllib2\n", | |
| "import json\n", | |
| "from StringIO import StringIO\n", | |
| "\n", | |
| "# custom code to display tweets inline by ID -allenday\n", | |
| "IPython.display.display_html('<script type=\"text/javascript\" src=\"//platform.twitter.com/widgets.js\"></script>',raw=True)\n", | |
| "def display_tweet(id):\n", | |
| " payload = urllib2.urlopen('https://api.twitter.com/1/statuses/oembed.json?id=%s' % id).read()\n", | |
| " payload_io = StringIO(payload)\n", | |
| " payload_json = json.load(payload_io)\n", | |
| " IPython.display.display_html(payload_json['html'],raw=True)\n", | |
| " \n", | |
| "IPython.display.YouTubeVideo(\"M3O5YRS4_kY\")" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "... and this excellent talk, [Add ALL The Things](http://www.infoq.com/presentations/abstract-algebra-analytics) by Avi Bryant @ Stripe." | |
| ] | |
| }, | |
| { | |
| "cell_type": "heading", | |
| "level": 2, | |
| "metadata": {}, | |
| "source": [ | |
| "Data Preparation" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "We'll need plenty of interesting data to illustrate how to use these kinds of algorithms... Let's create a data set from the text of the [Jabberwocky poem](http://www.jabberwocky.com/carroll/jabber/jabberwocky.html) by Lewis Carroll:" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": false, | |
| "input": [ | |
| "import re\n", | |
| "\n", | |
| "def clean_words (text):\n", | |
| " return filter(lambda x: len(x) > 0, re.sub(\"[^A-Za-z]\", \" \", text).split(\" \"))\n", | |
| "\n", | |
| "def clean_lines (text):\n", | |
| " return map(lambda x: clean_words(x), filter(lambda x: len(x) > 0, text.split(\"\\n\")))" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": false, | |
| "input": [ | |
| "jabber_text = \"\"\"\n", | |
| "`Twas brillig, and the slithy toves\n", | |
| " Did gyre and gimble in the wabe:\n", | |
| "All mimsy were the borogoves,\n", | |
| " And the mome raths outgrabe.\n", | |
| "\n", | |
| "\"Beware the Jabberwock, my son!\n", | |
| " The jaws that bite, the claws that catch!\n", | |
| "Beware the Jubjub bird, and shun\n", | |
| " The frumious Bandersnatch!\"\n", | |
| "He took his vorpal sword in hand:\n", | |
| " Long time the manxome foe he sought --\n", | |
| "So rested he by the Tumtum tree,\n", | |
| " And stood awhile in thought.\n", | |
| "And, as in uffish thought he stood,\n", | |
| " The Jabberwock, with eyes of flame,\n", | |
| "Came whiffling through the tulgey wood,\n", | |
| " And burbled as it came!\n", | |
| "One, two! One, two! And through and through\n", | |
| " The vorpal blade went snicker-snack!\n", | |
| "He left it dead, and with its head\n", | |
| " He went galumphing back.\n", | |
| "\"And, has thou slain the Jabberwock?\n", | |
| " Come to my arms, my beamish boy!\n", | |
| "O frabjous day! Callooh! Callay!'\n", | |
| " He chortled in his joy.\n", | |
| "\n", | |
| "`Twas brillig, and the slithy toves\n", | |
| " Did gyre and gimble in the wabe;\n", | |
| "All mimsy were the borogoves,\n", | |
| " And the mome raths outgrabe.\n", | |
| "\"\"\"\n", | |
| "\n", | |
| "jabber_words = clean_words(jabber_text.lower())\n", | |
| "print \"full:\", jabber_words\n", | |
| "\n", | |
| "print \"\\n*****\\n\"\n", | |
| "\n", | |
| "jabber_uniq = sorted(set(jabber_words))\n", | |
| "print \"uniq:\", jabber_uniq\n", | |
| "\n", | |
| "print \"\\n*****\\n\"\n", | |
| "\n", | |
| "jabber_line = clean_lines(jabber_text.lower())\n", | |
| "print \"sentences:\", jabber_line" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "heading", | |
| "level": 3, | |
| "metadata": {}, | |
| "source": [ | |
| "Algorithm: HyperLogLog" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "**Problem:** Now let's use that data set with [HyperLogLog](http://en.wikipedia.org/wiki/HyperLogLog) to implement a probabilistic cardinality counter.\n", | |
| "\n", | |
| "Let's start with an interactive demo of the HyperLogLog algorithm:" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": false, | |
| "input": [ | |
| "display_tweet(519102279710806016)" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Implementation" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": false, | |
| "input": [ | |
| "import hyperloglog\n", | |
| "\n", | |
| "# accept 1% counting error\n", | |
| "hll = hyperloglog.HyperLogLog(0.01)\n", | |
| "\n", | |
| "for word in jabber_words:\n", | |
| " hll.add(word)\n", | |
| "\n", | |
| "print \"prob count %d, true count %d\" % (len(hll), len(jabber_uniq))\n", | |
| "print \"observed error rate %0.2f\" % (abs(len(hll) - len(jabber_uniq)) / float(len(jabber_uniq)))" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Great, those results show how the bounded error rate is working as expected." | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "By the way, [http://research.neustar.biz](http://research.neustar.biz) is an all around excellent resouce for learning about approximation algorithms and probabilistic data structures. Check it out." | |
| ] | |
| }, | |
| { | |
| "cell_type": "heading", | |
| "level": 3, | |
| "metadata": {}, | |
| "source": [ | |
| "Algorithm: Count-Min Sketch" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "**Problem:** Next we'll use [Count-min sketch](http://en.wikipedia.org/wiki/Count%E2%80%93min_sketch) for frequency summaries, to implement a probabilistic [word count](http://en.wikipedia.org/wiki/Word_count)...\n", | |
| "\n", | |
| "There's also descriptions and figures of Count-Min sketch [here](http://lkozma.net/blog/sketching-data-structures/) and [here](http://research.neustar.biz/tag/count-min-sketch/)" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": false, | |
| "input": [ | |
| "from countminsketch import CountMinSketch\n", | |
| "\n", | |
| "# table size=1000, hash functions=10\n", | |
| "cms = CountMinSketch(1000, 10)\n", | |
| "\n", | |
| "# add the word to the count-min sketch\n", | |
| "for word in jabber_words:\n", | |
| " cms.add(word)\n", | |
| "\n", | |
| "### reporting\n", | |
| "# get the probable word counts for each word\n", | |
| "pwc = [(word, cms[word],) for word in jabber_uniq]\n", | |
| "\n", | |
| "# get the true word counts for each word\n", | |
| "twc = {}\n", | |
| "for word in jabber_words:\n", | |
| " if (word in twc):\n", | |
| " twc[word] += 1\n", | |
| " else:\n", | |
| " twc[word] = 1\n", | |
| "\n", | |
| "# compare and pretty-print the probable and true word counts\n", | |
| "from operator import itemgetter\n", | |
| "\n", | |
| "# show error and count of countminsketch\n", | |
| "e = list()\n", | |
| "for pair in sorted(pwc, key=itemgetter(1), reverse=True):\n", | |
| " e.append(pair[1] - twc[pair[0]])\n", | |
| " print \"d=%s %s\" % (abs(pair[1] - twc[pair[0]]), pair)\n", | |
| "print e" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Try reducing table_size to 20 and get data like this:" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": false, | |
| "input": [ | |
| "[2, 0, 3, 1, 4, 3, 5, 3, 5, 4, 4, 3, 4, 4, 4, 3, 4, 4, 4, 4, 3, 4, 4, 4, 4, 3, 3, 2, 3, 3, 2, 2, 3, 3, 3, 3, 3, 3, 2, 3, 2, 2, 2, 3, 3, 2, 2, 1, 3, 2, 1, 2, 2, 2, 2, 2, 2, 1, 2, 1, 2, 2, 2, 2, 2, 1, 2, 2, 2, 2, 2, 1, 2, 1, 1, 2, 2, 1, 1, 1, 1, 0, 1, 1, 0, 0, 1, 1, 0, 0, 0]" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "**Problem:** why?" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Bokay, that was hella simpler than writting 2000 lines of Java based on Yet-Another-Hadoop-Nightmare (YAHN)." | |
| ] | |
| }, | |
| { | |
| "cell_type": "heading", | |
| "level": 3, | |
| "metadata": {}, | |
| "source": [ | |
| "Algorithm: Bloom Filter" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "**Problem:** Next, let's use a [Bloom filter](http://en.wikipedia.org/wiki/Bloom_filter) for approximate membership queries... In this case, we'll compare text from a local markdown file `INSTALL.md` to see if it has words intersecting with *Jabberwocky*?\n", | |
| "\n", | |
| "Here's an interactive demo of the Bloom Filter:" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": false, | |
| "input": [ | |
| "display_tweet(522030172263186434)" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": false, | |
| "input": [ | |
| "from pybloom import BloomFilter\n", | |
| "\n", | |
| "bf = BloomFilter(capacity=1000, error_rate=0.001)" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": false, | |
| "input": [ | |
| "for word in jabber_words:\n", | |
| " bf.add(word)\n", | |
| "\n", | |
| "intersect = set([])\n", | |
| "\n", | |
| "with open(\"INSTALL.md\") as f:\n", | |
| " for line in f:\n", | |
| " for word in clean_words(line.strip().lower()):\n", | |
| " if word in bf:\n", | |
| " intersect.add(word)\n", | |
| "\n", | |
| "print intersect" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "heading", | |
| "level": 2, | |
| "metadata": {}, | |
| "source": [ | |
| "Combining Algorithms" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Now let's take a look at how we can combine some probabilistic data structures/algorithms to build a useful application.\n", | |
| "\n", | |
| "This example is based on an excellent article _Multi-Index Locality Sensitive Hashing for Fun and Profit_ from [Jay Chan](https://github.com/justecorruptio) of [EventBrite](http://eventbrite.com) found [here](https://engineering.eventbrite.com/multi-index-locality-sensitive-hashing-for-fun-and-profit/).\n", | |
| "\n", | |
| "For any line in the Jabberwock poem, we'll build a system for retrieving similar lines **efficiently** with regard to CPU cost, but at increased storage cost. We'll show how to use probabilistic data structures to refactor this problem (retrieve similar lines).\n", | |
| "\n", | |
| "The _naive_ implementation has computational complexity **O(lines^2 words^2)**\n", | |
| "\n", | |
| "The _probabilistic data structures_ has an implementation that has computational complexity **O(lines)**" | |
| ] | |
| }, | |
| { | |
| "cell_type": "heading", | |
| "level": 3, | |
| "metadata": {}, | |
| "source": [ | |
| "A Brief Digression: The Jaccard Similarity Coefficient" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Frequently probabilistic data structures are used when an application requires comparing objects, (e.g. sorting, clustering), but there are simply too many objects to keep in memory or the comparison operation takes too long.\n", | |
| "\n", | |
| "As a concrete example of object comparison, let's look at the [Jaccard Similarity Coefficient](http://en.wikipedia.org/wiki/Jaccard_index) which defines a scheme for calculating the distance between two sets (A,B) that is bounded 0..1. Mathematically we can describe it as the set intersection divided by the set union, like this:\n", | |
| "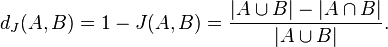" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": false, | |
| "input": [ | |
| "from sets import Set\n", | |
| "\n", | |
| "def L2S (X):\n", | |
| " A = Set()\n", | |
| " for x in X:\n", | |
| " A.add(x)\n", | |
| " return A\n", | |
| "\n", | |
| "def jaccard_similarity (A, B):\n", | |
| " i = A.intersection(B)\n", | |
| " u = A.union(B)\n", | |
| " return(float(len(i))/len(u))" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Now let's see it in action by taking a copule of lines and measuring their similarity" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": false, | |
| "input": [ | |
| "s0 = jabber_line[0]\n", | |
| "s1 = jabber_line[1]\n", | |
| "s24 = jabber_line[24]\n", | |
| "\n", | |
| "print \"line0: \", s0\n", | |
| "print \"line1: \", s1\n", | |
| "print \"line24: \", s24\n", | |
| "print \"J(0,1): \", jaccard_similarity(L2S(s0), L2S(s1))\n", | |
| "print \"J(0,24): \", jaccard_similarity(L2S(s0), L2S(s24))" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "One problem with Jaccard similarity is that it is affected by set size. Because the coefficient's denominator is the size of the union of all words, it will have a smaller value when one or both sets have many items. One way around this problem is to use another probabilistic data structure: the MinHash.\n", | |
| "\n", | |
| "Another problem is the computational complexity to use Jaccard.\n", | |
| "\n", | |
| "To compare a pair of lines, it costs **O(wordsA * wordsB)** operations, or mean **O(words^2)** across the whole dataset. This, combined with comparing all pairs of lines gives a computational complexity of **O(words^2 lines^2)**. Not good." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": false, | |
| "input": [ | |
| "IPython.display.Image(\"http://i.imgur.com/iU6wQvv.jpg\")" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "heading", | |
| "level": 3, | |
| "metadata": {}, | |
| "source": [ | |
| "Algorithm: MinHash" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "It requires defining a # of hash functions which we'll call **H**\n", | |
| "\n", | |
| "MinHash implementation has a few benefits over Jaccard:\n", | |
| "1. improved distance - distance scale is no longer affected by the cardinality (size) of either set because the # of hash functions **H** is constant.\n", | |
| "2. improved storage requirements - we can store **H** numbers (short, int, long, etc) rather than keeping all words in the indexing system. However, note that this does not affect storage requirements for the original documents, should that be necessary.\n", | |
| "3. improved time complexity - rather than needing to compare all items in set A vs all items in set B (naively **O(words^2)** and at best **O(words log words)**), we can perform a fixed **H** comparisons." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": false, | |
| "input": [ | |
| "import md5\n", | |
| "from struct import unpack\n", | |
| "\n", | |
| "# H is selected empirically based on desired sensitivity/complexity tradeoff\n", | |
| "H = 100\n", | |
| "\n", | |
| "def hash_func (text):\n", | |
| " return unpack(\"i\",str((md5.md5(text).digest())[0:4]))\n", | |
| "\n", | |
| "# words is the set of words previously defined\n", | |
| "# returns a vector of N hashes\n", | |
| "def minhash (words, N):\n", | |
| " # number of hashes to calculate\n", | |
| " hashes = list()\n", | |
| " for seed in range(0, N):\n", | |
| " hash_list = list()\n", | |
| " for word in words:\n", | |
| " #hash function returns an int\n", | |
| " hash_val = hash_func(str(seed)+word)\n", | |
| " hash_list.append(hash_val[0])\n", | |
| " min_hash = min(hash_list)\n", | |
| " hashes.append(min_hash)\n", | |
| " return hashes\n", | |
| " \n", | |
| "\n", | |
| "\n", | |
| "\n" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": false, | |
| "input": [ | |
| "def minhash_similarity (textA, textB, N):\n", | |
| " vA = minhash(textA, N)\n", | |
| " vB = minhash(textB, N)\n", | |
| " count = 0\n", | |
| " for i in range(0, N):\n", | |
| " if vA[i] == vB[i]:\n", | |
| " count += 1\n", | |
| " return float(count) / N\n", | |
| " \n", | |
| "print minhash(s0, H)\n", | |
| "print minhash_similarity(s0, s24, H)\n", | |
| "print minhash_similarity(s0, s1, H)\n", | |
| "\n", | |
| "print \"\\n*****\\n\"\n", | |
| "\n", | |
| "for i in range(0,len(jabber_line)):\n", | |
| " for j in range(0,len(jabber_line)):\n", | |
| " if i < j:\n", | |
| " si = jabber_line[i]\n", | |
| " sj = jabber_line[j]\n", | |
| " sim = minhash_similarity(si, sj, H)\n", | |
| " if sim > 0.5:\n", | |
| " print (sim*100), \"% similar lines:\\n [\",i,\"]\", si, \"\\n [\",j,\"]\", sj\n" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "It's an improvement over the Jaccard implementation in that we factored the **O(words^2)** component down to **O(H)**, for a total complexity of now **O(lines^2 H)**. To reduce this lookup cost, we introduce new costs of **S = storage** and **I = indexing** of **O(H lines)** and **H lines** respectively." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": false, | |
| "input": [ | |
| "IPython.display.Image(\"http://i.imgur.com/IQUhP0c.jpg\")" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "We can take this a step further and eliminate the H component using a technique called [bit sampling](http://en.wikipedia.org/wiki/Locality-sensitive_hashing#Bit_sampling_for_Hamming_distance).\n", | |
| "\n", | |
| "The idea here is to take the least significant bit of each hash, and keep those H bits as a new, single bit vector that represents the input. Note that the distance cutoff we use to consider two lines as close needs to increase, because the LSB will have the same value in two unrelated lines 50% of the time.\n", | |
| "\n", | |
| "**Problem**: define cutoff for similarity as function of H. This relates to the [binomial distribution](http://en.wikipedia.org/wiki/Binomial_distribution) and is left as an exercise for the avid student.\n", | |
| "\n", | |
| "While we introduce a noisy distance estimator, we achieve some gain also. Namely the storage required is reduced linearly from **H integers** to **H**. Also, the comparison between any pair of lines is reduced to approximately **O(1)** for reasonable values of **H** because the bit comparisons can be made in parallel in a CPU or GPU (eg 32 bits, 64 bits)." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": false, | |
| "input": [ | |
| "IPython.display.Image(\"http://i.imgur.com/4OVHpl0.jpg\")" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Here's the code." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": false, | |
| "input": [ | |
| "def bitsample(hashes, N):\n", | |
| " # returns bits, which is a bit vector of the least significant bits\n", | |
| " # of all the hashes. This will have to span multiple integers for large N.\n", | |
| " bits = 0\n", | |
| " \n", | |
| " for i in range(0, N):\n", | |
| " hash = hashes[i]\n", | |
| " #print(hash)\n", | |
| " bits = (bits << 1) | (hash & 1)\n", | |
| " return bits\n", | |
| "\n", | |
| "def hamming(bA, bB, N): \n", | |
| " X = bin(bA ^ bB)[2:] #xor bits\n", | |
| " while len(X) < N: #pad with leading zeroes\n", | |
| " X = \"0\" + X\n", | |
| " count = 0\n", | |
| " # this can be reduced to O(1).\n", | |
| " # see: http://graphics.stanford.edu/~seander/bithacks.html#CountBitsSetParallel\n", | |
| " for i in range(0, N):\n", | |
| " if int(X[i:i+1]) == 1:\n", | |
| " count += 1 \n", | |
| " return count\n", | |
| " \n", | |
| "def bit_similarity(textA, textB, N):\n", | |
| " bitsA = bitsample(minhash(textA,H),N)\n", | |
| " bitsB = bitsample(minhash(textB,H),N)\n", | |
| " distance = hamming(bitsA, bitsB, N)\n", | |
| " sim = 1 - (float(distance) / N)\n", | |
| " return sim\n", | |
| "\n", | |
| "b0 = bitsample(minhash(s0,H),H)\n", | |
| "b1 = bitsample(minhash(s1,H),H)\n", | |
| "\n", | |
| "print s0\n", | |
| "print jabber_line[0]\n", | |
| "print \"s0 \", bin(b0)[2:]\n", | |
| "print \"s1 \", bin(b1)[2:]\n", | |
| "print \"B(s0, s1) \",bit_similarity(s0, s1, H)\n", | |
| "\n", | |
| "print \"\\n*****\\n\"\n", | |
| "\n", | |
| "for i in range(0,len(jabber_line)):\n", | |
| "# print bitsample(minhash(jabber_line[i],H),H)\n", | |
| " for j in range(0,len(jabber_line)):\n", | |
| " if i < j:\n", | |
| " si = jabber_line[i]\n", | |
| " sj = jabber_line[j]\n", | |
| " sim = bit_similarity(si, sj, H)\n", | |
| " if sim > 0.7:\n", | |
| " print (sim*100), \"% similar sentences:\\n [\",i,\"]\", si, \"\\n [\",j,\"]\", sj\n" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "It's an improvement over the Jaccard implementation in that we factored the **O(words^2)** component down to **O(H)**, for a total complexity of now **O(lines^2 H)**. To reduce this lookup cost, we introduce new costs of **S = storage = H lines** and **I = indexing = O(H lines)**.\n", | |
| "\n", | |
| "However, we're still paying **O(lines^2)** for comparison. Onward." | |
| ] | |
| }, | |
| { | |
| "cell_type": "heading", | |
| "level": 3, | |
| "metadata": {}, | |
| "source": [ | |
| "Locality-Sensitive Hashing" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "In the common case, hashing functions are used to distribute a set of inputs uniformly across the range of the hash space. This is good for e.g. achieving uniform workload across a distributed system and as such using a hash inside of HBase/NoSQL keys is considered a [best practice](http://hbase.apache.org/book/rowkey.design.html).\n", | |
| "\n", | |
| "Sometimes however, it's desireable to have a hashing function that puts similar inputs nearby to one another in the hash space. This is called [locality-sensitive hashing](http://en.wikipedia.org/wiki/Locality-sensitive_hashing) and is analagous to the linear algebra / machine learning technique of dimension reduction (e.g. [eigenvectors](http://en.wikipedia.org/wiki/Eigenvalues_and_eigenvectors), [principal components analysis](http://en.wikipedia.org/wiki/Principal_component_analysis).\n", | |
| "\n", | |
| "Given that data can only be stored in one primary order in computer address space, this presents a problem for K-dimensional data. So, usually you see this technique show up for data points that are inherently K-dimensional, and you want to be able to look them up by their position (and find neighbors) in that K-dimensional space.\n", | |
| "\n", | |
| "Here's an interactive demo of a type of LSH for geospatial (K=2) data called [geohashing](http://en.wikipedia.org/wiki/Geohash)." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": false, | |
| "input": [ | |
| "display_tweet(\"522375375952044032\")" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "We'll do something similar for our bitsample vector.\n", | |
| "\n", | |
| "Remember, each of the bits is originally derived from one of the minhash values, and these bits/minhashes can be considered as independent dimensions of the input lines.\n", | |
| "\n", | |
| "As an aside, one (older) data structure for addressing this kind of problem is a [KD-Tree](http://en.wikipedia.org/wiki/K-d_tree).\n", | |
| "\n", | |
| "We'll look at a (newer) technique that can produce similar results at lower cost called [multi-index LSH](http://scholar.google.com/scholar?espv=2&bav=on.2,or.r_cp.r_qf.&bvm=bv.77412846,d.eXY&ion=1&biw=1345&bih=679&um=1&ie=UTF-8&lr=&cites=3413868449671240253).\n", | |
| "\n", | |
| "The advantage of the latter technique is that it scales linearly, whereas the KD-Tree has familiar **O(n log n)** scaling characteristics common to [other trees you may know](http://en.wikipedia.org/wiki/Tree_(graph_theory)#rooted_tree)." | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": false, | |
| "input": [ | |
| "class LshTable: \n", | |
| " def __init__(self, _N, _K):\n", | |
| " self.N = _N #bits in vector\n", | |
| " self.K = _K #chunks of bits\n", | |
| " self.C = _N/_K\n", | |
| " self.hash_tables = list()\n", | |
| " #R = radius - not implemented\n", | |
| " \n", | |
| " for i in range(0, self.K):\n", | |
| " self.hash_tables.append({})\n", | |
| "\n", | |
| " def generate_close_chunks(self, chunk):\n", | |
| " # returns a list of all possible chunks that is zero or one bit off of the input\n", | |
| " # eg: find_close_chunks(0000) -> [0000, 1000, 0100, 0010, 0001]\n", | |
| " # eg: find_close_chunks(1011) -> [1011, 0011, 0111, 1001, 1010]\n", | |
| " X = bin(chunk)[2:]\n", | |
| " while len(X) < self.C: #pad with leading zeroes\n", | |
| " X = \"0\" + X\n", | |
| "\n", | |
| " close_chunks = list()\n", | |
| " close_chunks.append(chunk)\n", | |
| "\n", | |
| " for i in range(1, self.C):\n", | |
| " T = X[:i]\n", | |
| " if X[i] == \"1\":\n", | |
| " T = T + \"0\"\n", | |
| " else:\n", | |
| " T = T + \"1\"\n", | |
| " T = T + X[i+1:]\n", | |
| " close_chunks.append(int(T,2)) \n", | |
| " return close_chunks\n", | |
| " \n", | |
| " def split_chunks(self, v):\n", | |
| " # splits a bit vector into K chunks\n", | |
| " X = bin(v)[2:]\n", | |
| " c = len(X)/self.K\n", | |
| " chunks = list()\n", | |
| " for i in range(0, self.K):\n", | |
| " chunks.append(int(X[(i*c):(i*c)+c],2))\n", | |
| " return chunks\n", | |
| " \n", | |
| " def add(self, bit_vector):\n", | |
| " chunks = self.split_chunks(bit_vector)\n", | |
| " \n", | |
| " for i in range(1, self.K):\n", | |
| " hash_table = self.hash_tables[i] # fetch the ith hash table\n", | |
| " chunk = chunks[i] # fetch the ith chunk of the bit_vector\n", | |
| " \n", | |
| " if chunk not in hash_table:\n", | |
| " hash_table[chunk] = list()\n", | |
| " \n", | |
| " bit_vector_list = hash_table[chunk]\n", | |
| " bit_vector_list.append(bit_vector)\n", | |
| " \n", | |
| " def lookup(self, query, cutoff):\n", | |
| " chunks = self.split_chunks(query)\n", | |
| " \n", | |
| " possible_matches = {}\n", | |
| " for i in range(1, self.K):\n", | |
| " hash_table = self.hash_tables[i]\n", | |
| " chunk = chunks[i]\n", | |
| " \n", | |
| " close_chunks = self.generate_close_chunks(chunk)\n", | |
| " \n", | |
| " for close_chunk in close_chunks:\n", | |
| " if close_chunk in hash_table:\n", | |
| " for match in hash_table[close_chunk]:\n", | |
| " possible_matches[match] = 1\n", | |
| " \n", | |
| " verified_matches = {}\n", | |
| " for match in possible_matches:\n", | |
| " distance = hamming(query, match, self.N)\n", | |
| " sim = 1 - (float(distance) / self.N)\n", | |
| " if sim > cutoff:\n", | |
| " verified_matches[match] = sim\n", | |
| " \n", | |
| " return verified_matches\n", | |
| "\n", | |
| "K = 25\n", | |
| "t = LshTable(H,K)\n", | |
| "#print t.generate_close_chunks(15)\n", | |
| "#print t.generate_close_chunks(8)\n", | |
| "#print bin(b0)\n", | |
| "#print t.split_chunks(b0)\n", | |
| "\n", | |
| "# book-keeping for viz later. *yawn*\n", | |
| "bitsample_lineid = {}\n", | |
| "lineid_line = {}\n", | |
| "i = 0\n", | |
| "for line in jabber_line:\n", | |
| " bs = bitsample(minhash(line,H),H)\n", | |
| " bitsample_lineid[bs] = i #line\n", | |
| " lineid_line[i] = line\n", | |
| " t.add(bs)\n", | |
| " i += 1\n", | |
| "\n", | |
| "i = 0\n", | |
| "for line in jabber_line:\n", | |
| " bs = bitsample(minhash(line,H),H)\n", | |
| " hits = t.lookup(bs, 0.7)\n", | |
| " if len(hits) > 0:\n", | |
| " print \"line\", i, line\n", | |
| " for hit in hits:\n", | |
| " if bitsample_lineid[hit] != i:\n", | |
| " print \" hit\",bitsample_lineid[hit],\"(\",hits[hit],\"):\", lineid_line[bitsample_lineid[hit]]\n", | |
| " i += 1\n", | |
| "\n", | |
| "# NB: identical line pairs (e.g. lines 0 and 24) don't find one another\n", | |
| "# this is because of the lookup hashes used for illustrative purposes\n", | |
| "# A fix is left as an exercise for the avid student\n", | |
| "\n", | |
| "#print t.lookup(bitsample(minhash(jabber_sent[3],H),H),0.6)\n", | |
| "#print t.lookup(bitsample(minhash(jabber_sent[27],H),H),0.6) \n", | |
| " \n", | |
| "#print bitsample(minhash(s0,H),H)\n", | |
| "#print \"query: \", s0\n", | |
| "#hits = t.lookup(b0, 0.5)\n", | |
| "#for hit in hits:\n", | |
| "# print \" hit (\",hits[hit],\"):\", bitsample_sent[hit]\n", | |
| "#print t.lookup(b0, 0.5)" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "Alright, check it out. We've now reduced the complexity of the lookup operation to **O(chunks)** for a single line. This is a big deal, because it means the overall lookup complexity is **O(lines chunks)**. As discussed earlier **chunks** is proportional to **H** and is performed in-parallel, in-register so the complexity is really **O(lines)**.\n", | |
| "\n", | |
| "This comes at additional indexing cost of **O(H lines)** (again, effectively **O(lines)**) and storage cost of **H chunks R**. Note that I just slipped in the **R** term -- this is the radius parameter, and allows lookup of items in the LSH index that are R bits difference from the query. For the implementation above **R=1**" | |
| ] | |
| }, | |
| { | |
| "cell_type": "code", | |
| "collapsed": false, | |
| "input": [ | |
| "IPython.display.Image(\"http://i.imgur.com/rHoahwv.jpg\")" | |
| ], | |
| "language": "python", | |
| "metadata": {}, | |
| "outputs": [] | |
| }, | |
| { | |
| "cell_type": "heading", | |
| "level": 2, | |
| "metadata": {}, | |
| "source": [ | |
| "Summary" | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "This is just a start. Even so, not so many programmers can claim that they have written and run code that takes advantage of probabilistic data structures. You have." | |
| ] | |
| }, | |
| { | |
| "cell_type": "markdown", | |
| "metadata": {}, | |
| "source": [ | |
| "*Installation notes:*\n", | |
| "\n", | |
| " * [HyperLogLog](https://github.com/svpcom/hyperloglog)\n", | |
| " * [CountMinSketch](https://github.com/rafacarrascosa/countminsketch)\n", | |
| " * [pybloom](https://pypi.python.org/pypi/pybloom/1.0.2)" | |
| ] | |
| } | |
| ], | |
| "metadata": {} | |
| } | |
| ] | |
| } |
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment