Last active
July 29, 2018 21:15
-
-
Save conjugateprior/1b1e8200af77948b33f05a0282807494 to your computer and use it in GitHub Desktop.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| ```{r} | |
| library(MonetDBLite) | |
| library(RSQLite) | |
| library(DBI) | |
| library(dplyr) | |
| library(microbenchmark) | |
| library(ggplot2) | |
| ``` | |
| Load 10M international events. This is a tidy version of the events | |
| downloadable from https://dataverse.harvard.edu/dataset.xhtml?persistentId=hdl:1902.1/FYXLAWZRIA ([King and Lowe, 2003](http://dx.doi.org/10.1017/S0020818303573064)). | |
| Size as a single `.rda` file on disk: *96.5M* | |
| ```{r} | |
| load("events-1990-2004.rda") | |
| # adjust column names for easier SQL import | |
| names(events) <- c("id", "xid", "sid", "eid", "place", | |
| "eventdate", "eventform", "srcname", | |
| "srcsector", "srclevel", "tgtname", | |
| "tgtsector", "tgtlevel") | |
| ``` | |
| Set up a Monet database in a local directory, make events a table, | |
| and create a table front for dplyr | |
| ```{r} | |
| dbdir <- "monet-dplyr-dir" | |
| con1 <- dbConnect(MonetDBLite::MonetDBLite(), dbdir) | |
| dbWriteTable(con1, "events", events, overwrite = TRUE) | |
| ev1 <- tbl(con1, "events") | |
| ``` | |
| Size as a folder on disk: *425M* | |
| Set up an SQLite database in a local directory, make events a table and | |
| create a table front for dplyr | |
| ```{r} | |
| dbfile <- "sqlite-dplyr.db" | |
| con2 <- dbConnect(RSQLite::SQLite(), dbfile) | |
| dbWriteTable(con2, "events", events, overwrite = TRUE) | |
| ev2 <- tbl(con2, "events") | |
| ``` | |
| Size as a single sqlite file on disk: *690M* | |
| Set up an SQLite database in memory and create a table from for dplyr | |
| ```{r} | |
| con3 <- dbConnect(RSQLite::SQLite(), ":memory:") | |
| dbWriteTable(con3, "events", events, overwrite = TRUE) | |
| ev3 <- tbl(con3, "events") | |
| ``` | |
| Set up an SQLite database in memory and create a table from for dplyr | |
| ```{r} | |
| con4 <- dbConnect(RSQLite::SQLite(), ":memory:") | |
| copy_to(con4, events, "events", | |
| indexes = list("eventform")) | |
| ev4 <- tbl(con4, "events") | |
| ``` | |
| We'll test a single variable tabulation. Here are the test functions: | |
| ```{r} | |
| # Base R's version, using table | |
| timing_r <- function(ev){ | |
| table(ev$eventform) | |
| } | |
| # SQL via DBI version | |
| timing_dbi <- function(con){ | |
| rr <- dbSendQuery(con, "SELECT eventform, COUNT(eventform) FROM events GROUP BY eventform") | |
| res <- dbFetch(rr) | |
| dbClearResult(rr) | |
| } | |
| # dplyr version | |
| timing_dplyr <- function(ev){ | |
| ev %>% | |
| select(eventform) %>% | |
| group_by(eventform) %>% | |
| summarise(count = n()) %>% | |
| collect() | |
| } | |
| ``` | |
| Run the test. This takes about 12 minutes on my laptop | |
| ```{r} | |
| res <- microbenchmark(r = timing_r(events), | |
| dbi1 = timing_dbi(con1), | |
| dbi2 = timing_dbi(con2), | |
| dbi3 = timing_dbi(con3), | |
| dbi4 = timing_dbi(con4), | |
| dplyrr = timing_dplyr(events), | |
| dplyr1 = timing_dplyr(ev1), | |
| dplyr2 = timing_dplyr(ev2), | |
| dplyr3 = timing_dplyr(ev3), | |
| dplyr4 = timing_dplyr(ev4), | |
| times = 30) | |
| ``` | |
| and close up the connections now we're done. | |
| ```{r} | |
| dbDisconnect(con1, shutdown = TRUE) | |
| dbDisconnect(con2, shutdown = TRUE) | |
| dbDisconnect(con3, shutdown = TRUE) | |
| dbDisconnect(con4, shutdown = TRUE) | |
| ``` | |
| Results: | |
| ```{r} | |
| tab <- summary(res) %>% | |
| mutate(library = case_when( | |
| grepl("dbi", expr) ~ "DBI", | |
| grepl("dplyr", expr) ~ "dplyr", | |
| expr == "r" ~ "base"), | |
| database = case_when( | |
| grepl("1", expr) ~ "Monet", | |
| grepl("2", expr) ~ "SQLite (disk)", | |
| grepl("3", expr) ~ "SQLite (memory)", | |
| grepl("4", expr) ~ "SQLite (memory+index)", | |
| TRUE ~ ""), | |
| seconds = time / 1000000000, | |
| Condition = paste(library, database)) | |
| ``` | |
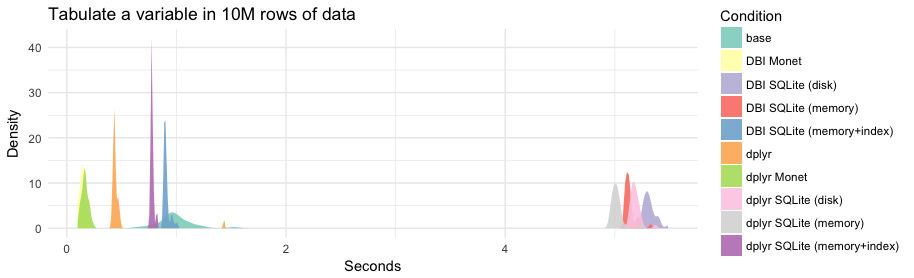
| Speed summaries (in milliseconds) | |
| | min| lq| mean| median| uq| max| neval|library |database | | |
| |----------:|---------:|---------:|---------:|---------:|---------:|-----:|:-------|:---------------------| | |
| | 654.82496| 908.8472| 990.2477| 970.7917| 1064.4448| 1526.8617| 30|base | | | |
| | 98.51502| 127.3214| 143.2917| 138.8620| 157.6495| 218.0188| 30|DBI |Monet | | |
| | 5227.39406| 5273.4724| 5315.1499| 5306.2525| 5338.7910| 5485.8420| 30|DBI |SQLite (disk) | | |
| | 5084.73081| 5101.5085| 5136.9485| 5123.0648| 5140.5585| 5326.1279| 30|DBI |SQLite (memory) | | |
| | 882.60921| 890.9514| 907.7341| 897.7100| 909.2202| 1003.5088| 30|DBI |SQLite (memory+index) | | |
| | 413.31117| 429.5574| 473.0778| 436.2304| 445.9132| 1433.8018| 30|dplyr | | | |
| | 113.06272| 150.7392| 165.2783| 164.3106| 187.6424| 237.1465| 30|dplyr |Monet | | |
| | 5127.23870| 5156.3244| 5186.5221| 5178.9403| 5206.6186| 5369.7247| 30|dplyr |SQLite (disk) | | |
| | 4949.46828| 4988.3494| 5016.4259| 5010.3839| 5034.5425| 5235.0919| 30|dplyr |SQLite (memory) | | |
| | 762.09109| 768.8625| 778.1123| 774.8939| 782.2196| 823.0093| 30|dplyr |SQLite (memory+index) | | |
| ```{r} | |
| theme_set(theme_minimal()) | |
| ggplot(tablong, aes(x = seconds, fill = Condition)) + | |
| geom_density(alpha = 0.9, col = NA) + | |
| scale_fill_brewer(palette = "Set3") + | |
| labs(title = "Tabulate a variable in 10M rows of data", | |
| x = "Seconds", y = "Density") | |
| ``` | |
|  |
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
OK some extras. What about
data.table?Well, I don't really know what I'm doing with
data.tablebut it seems like the test function should be something likeand if that's reasonable usage then the
R,dplyr, anddata.tabletimings come out likeSo
data.tableis competitive (maybe a tad slower than) Monet, when accessed by DBI.