-
-
Save gadenbuie/c83e078bf8c81b035e32c3fc0cf04ee8 to your computer and use it in GitHub Desktop.
| #' Render Table of Contents | |
| #' | |
| #' A simple function to extract headers from an RMarkdown or Markdown document | |
| #' and build a table of contents. Returns a markdown list with links to the | |
| #' headers using | |
| #' [pandoc header identifiers](http://pandoc.org/MANUAL.html#header-identifiers). | |
| #' | |
| #' WARNING: This function only works with hash-tag headers. | |
| #' | |
| #' Because this function returns only the markdown list, the header for the | |
| #' Table of Contents itself must be manually included in the text. Use | |
| #' `toc_header_name` to exclude the table of contents header from the TOC, or | |
| #' set to `NULL` for it to be included. | |
| #' | |
| #' @section Usage: | |
| #' Just drop in a chunk where you want the toc to appear (set `echo=FALSE`): | |
| #' | |
| #' # Table of Contents | |
| #' | |
| #' ```{r echo=FALSE} | |
| #' render_toc("/path/to/the/file.Rmd") | |
| #' ``` | |
| #' | |
| #' @param filename Name of RMarkdown or Markdown document | |
| #' @param toc_header_name The table of contents header name. If specified, any | |
| #' header with this format will not be included in the TOC. Set to `NULL` to | |
| #' include the TOC itself in the TOC (but why?). | |
| #' @param base_level Starting level of the lowest header level. Any headers | |
| #' prior to the first header at the base_level are dropped silently. | |
| #' @param toc_depth Maximum depth for TOC, relative to base_level. Default is | |
| #' `toc_depth = 3`, which results in a TOC of at most 3 levels. | |
| render_toc <- function( | |
| filename, | |
| toc_header_name = "Table of Contents", | |
| base_level = NULL, | |
| toc_depth = 3 | |
| ) { | |
| x <- readLines(filename, warn = FALSE) | |
| x <- paste(x, collapse = "\n") | |
| x <- paste0("\n", x, "\n") | |
| for (i in 5:3) { | |
| regex_code_fence <- paste0("\n[`]{", i, "}.+?[`]{", i, "}\n") | |
| x <- gsub(regex_code_fence, "", x) | |
| } | |
| x <- strsplit(x, "\n")[[1]] | |
| x <- x[grepl("^#+", x)] | |
| if (!is.null(toc_header_name)) | |
| x <- x[!grepl(paste0("^#+ ", toc_header_name), x)] | |
| if (is.null(base_level)) | |
| base_level <- min(sapply(gsub("(#+).+", "\\1", x), nchar)) | |
| start_at_base_level <- FALSE | |
| x <- sapply(x, function(h) { | |
| level <- nchar(gsub("(#+).+", "\\1", h)) - base_level | |
| if (level < 0) { | |
| stop("Cannot have negative header levels. Problematic header \"", h, '" ', | |
| "was considered level ", level, ". Please adjust `base_level`.") | |
| } | |
| if (level > toc_depth - 1) return("") | |
| if (!start_at_base_level && level == 0) start_at_base_level <<- TRUE | |
| if (!start_at_base_level) return("") | |
| if (grepl("\\{#.+\\}(\\s+)?$", h)) { | |
| # has special header slug | |
| header_text <- gsub("#+ (.+)\\s+?\\{.+$", "\\1", h) | |
| header_slug <- gsub(".+\\{\\s?#([-_.a-zA-Z]+).+", "\\1", h) | |
| } else { | |
| header_text <- gsub("#+\\s+?", "", h) | |
| header_text <- gsub("\\s+?\\{.+\\}\\s*$", "", header_text) # strip { .tabset ... } | |
| header_text <- gsub("^[^[:alpha:]]*\\s*", "", header_text) # remove up to first alpha char | |
| header_slug <- paste(strsplit(header_text, " ")[[1]], collapse="-") | |
| header_slug <- tolower(header_slug) | |
| } | |
| paste0(strrep(" ", level * 4), "- [", header_text, "](#", header_slug, ")") | |
| }) | |
| x <- x[x != ""] | |
| knitr::asis_output(paste(x, collapse = "\n")) | |
| } |
| --- | |
| title: "blogdown toc example" | |
| author: '@gadenbuie' | |
| date: "2/28/2018" | |
| output: html_document | |
| --- | |
| ```{r setup, include=FALSE} | |
| knitr::opts_chunk$set(echo = TRUE) | |
| source("render_toc.R") | |
| ``` | |
| ## Table of Contents {#crazy-slug-here} | |
| ```{r toc, echo=FALSE} | |
| render_toc("blogdown-toc-example.Rmd") | |
| ``` | |
| ### WONT BE INCLUDED IN TOC | |
| # Writing | |
| ## R Markdown | |
| This is an R Markdown document. Markdown is a simple formatting syntax for authoring HTML, PDF, and MS Word documents. For more details on using R Markdown see <http://rmarkdown.rstudio.com>. | |
| When you click the **Knit** button a document will be generated that includes both content as well as the output of any embedded R code chunks within the document. You can embed an R code chunk like this: | |
| ```{r cars} | |
| # This is not a header | |
| summary(cars) | |
| ``` | |
| ## Regular Code | |
| ```r | |
| # Regular markdown code (not run) | |
| ggplot(mtcars, aes(x = wt, y = mpg)) + | |
| geom_point() | |
| ``` | |
| # Plots | |
| ## Including Plots {#plots-are-here .class-foo} | |
| You can also embed plots, for example: | |
| ```{r pressure, echo=FALSE} | |
| plot(pressure) | |
| ``` | |
| Note that the `echo = FALSE` parameter was added to the code chunk to prevent printing of the R code that generated the plot. | |
| # More | |
| ## Level 1 | |
| ### Level 1.a | |
| #### Level 1.a.i | |
| ### Level 1.b |
Thanks for the function. It works great and is very useful. However if one uses {.tabset .tabset-fade} for some of the headers, this will be included in the TOC generated by the function. Hopefully this is easy to fix.
Cheers
@JSkjoldahl Thanks! You're right, I forgot about those. I just added a fix for that that should strip any trailing {.class} declarations. Let me know if it works for you.
Hi @gadenbuie,
Thanks a lot for the function!
When I tried using it on my Rmarkdown file, the link target were not working.
I included in my titles some numeration, e.g.,
-
Step 1
-
Step 2
-
Step 3
By removing the pattern in line 67 of the initial code it worked out.header_slug <- paste(strsplit(gsub("[0-9]\\. ","",header_text), " ")[[1]], collapse="-")
I forked your function to edit it but can't figure out how to push back to you at the moment.
Leaving a comment instead in case it can help others...
Thanks again
render_toc() works to enable me to have the toc at any location on my rendered word doc. But still it doesnt help me to start the toc on a fresh page after the title page. The toc is rendered immediately after the title (see image below). Any ideas forcing toc onto a fresh page, short of inserting a page break manually after the word doc is rendered?

@rosalieb Thanks for letting me know! After a bit of thought, I remembered that pandoc makes sure that the HTML IDs start with a letter and removes anything that isn't a letter at the start of the ID. I updated the script to catch this. Cheers!
@bala-srm Short of creating a customized Word template for Pandoc, I think that manually inserting page breaks is your best option
Hi @gadenbuie,
Thank you for this great function, it's very useful!
As a question from a French user, is there any way to set up a different encoding for the table of contents (such as uft-8), so that all letters could be included?
Many thanks!
Hey @gadenbuie! Thank you so much for this. Just threw into an (incredibly long) post, and it worked like a charm 😄
Hello @gadenbuie, I am using this in a PDF and the standard TOC uses numbering and a bunch of formatting that is replaced with, for instance, bullet points and dashes when I change the location using this. Is there a way for it to retain the same format as before? Thanks!
Hello @gadenbuie, thanks for the function! I'm using the function to generate a TOC for several chapters in a bookdown project, but it doesn't seem to like the particular header:
#### What does 95% mean? {-}
It gives the error message:
Error in rawToChar(out) : embedded nul in string: '#what-does-95\0ean?' Calls: <Anonymous> ... <Anonymous> -> move_files_html -> vapply -> FUN -> rawToChar In addition: Warning message: In FUN(X[[i]], ...) : out-of-range values treated as 0 in coercion to raw
Would there be a reason why the function doesn't like the % symbol?
How can we have numbers instead of Bullet points and dashes.?
Hi @gadenbuie, thank you very much for the nice function, it worked perfectly if I don't use cat(). I have to use cat() in my report.
For example I add this chunk codes, it didn't recognize the heading in the table of content. Can you please help me to take a look?
You can see the table of contents only test 1, test 2, and test 3, setosa, versicolor, virginica did not include in table of content
Thank you very much
library(ggplot2)
for(Species in levels(iris$Species)){
cat('\n#', Species, '\n')
p <- ggplot(iris[iris$Species == Species,], aes(x = Sepal.Length, y = Sepal.Width)) +
geom_point()
print(p)
cat('\n')
}
I'm just here to support for the @Alchins question about encoding. I'm facing the same problem too. Tks.
@AugustoCL can you share an example where this function fails? I tried to create a problematic Rmd but was unable to reproduce the issue you and @Alchins describe.
I create this .Rmd with the topics which I got problems with encoding.
https://gist.github.com/AugustoCL/52b1f861c69e577463ef6c26cddfa820
The topics are in portuguese and it's very common have accents.
@AugustoCL I just tried that example and render_toc() worked as expected. Maybe you could try adjusting the encoding argument of readLines() on line 38?
I edit the encoding argument readLines() function, like you suggested and now it's working. Tks a lot for your help.
Many thanks for this. Trying to get rmarkdown html output into WCAG2.1-compliant format. This function is going to help in rebuilding the TOC for html output. I made a small adjustment:
knitr::asis_output(paste("<nav id=\"TOC\">",
paste(x, collapse = "\n"),
"</nav>", sep = "\n"))
I would like to credit you appropriately in the package docs, if you are happy for this.
Is there a way to enable automatic chapter numbering comparable to the suboption 'numbered_section=true' in the normal toc-command of the yaml-header? Instead of having a bullet-point list as the TOC, maybe is it possible to recreate this with a numbered list instead?
I can't test anything right now because my system is bricked, and this is beyond my capabilities anyways. However this would be a great addition to get rid of the pesky standard TOC...
Edit 1 17.08.2021 22:00:16:
I tested it on a friend's computer, and got some weird results. Aside from the fact that numbering doesn't seem to be possible (both automatic and in-header manual numbering, surprisingly, don't show up so far), the subsections of "Methods" are not listed beneath and indented to each other as they would be in a normal toc. I don't know why this is the case. I am running Win10, tested for word, pdf and html output respectively. Not sure why this is the case.

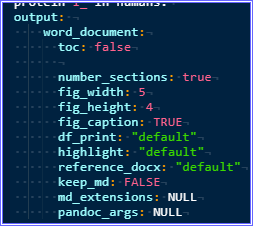
Is there a way to fix this, and possibly add a method of converting to numbered sections? Especially the first one is an absolute dealbreaker right now, but judging from other people's screenshots I might be encountering a bug or I am doing some stupid mistake I am not aware of.
Thank you for this gist.
It seems that blogdown is removing special characters from id in title sections, then !@:? need to be removed.
According to my test, I think this will do the trick:
h <- "éà@ù ?!: tot%" # expected: "[éà@ù ?!: tot%](éàù-tot)"
header_text <- gsub("#+\\s+?", "", h)
header_text <- gsub("\\s+?\\{.+\\}\\s*$", "", header_text) # strip { .tabset ... }
header_text <- gsub("^[^[:alpha:]]*\\s*", "", header_text) # remove up to first alpha char
# Remove special characters from slug
header_slug <- gsub("(\\W*)", "",
strsplit(header_text, " |-")[[1]] # keep `-` if exists
)
# Remove empty words
header_slug <- header_slug[header_slug != ""]
header_slug <- paste(header_slug, collapse = "-")
header_slug <- tolower(header_slug)Nice work! Wondering if there is anyway to change the color of the toc titles?
Hi, this is a very useful function. Thanks. But is there a way to keep the original chapter numbers instead of bullets?


Note: characters like
(,),/, ` etc are removed from the link target but this script doesn't take that into account