Created
January 10, 2022 07:08
-
-
Save linnil1/e6490fa7e8d44532b7a8eb2405206ffa to your computer and use it in GitHub Desktop.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| """ | |
| Airflow Example: Build a dynamic read-mapping DAG for multiple samples | |
| Author: linnil1 | |
| Run bowtie2(read-maper) on two samples to a reference fasta | |
| 1. Prepare data like this | |
| ``` | |
| $ ls /home/linnil1/airflow/data/ | |
| chrx1.synthetic.read1.fq chrx1.synthetic.read2.fq chrx.fa chrx.synthetic.read1.fq chrx.synthetic.read2.fq | |
| ``` | |
| 2. Setup your airflow | |
| https://github.com/linnil1/cwl_example#run-with-airflow | |
| 3. Setup variaribles (in Admin/variaribles) | |
| ``` | |
| basic_folder /home/linnil1/airflow/ | |
| reads data/chrx.synthetic,data/chrx1.synthetic | |
| reference_fa data/chrx.fa | |
| ``` | |
| 4. Move this file to airflow/dags | |
| 5. Trigger It | |
| """ | |
| from airflow import DAG | |
| from textwrap import dedent | |
| from datetime import datetime, timedelta | |
| from airflow.operators.bash import BashOperator | |
| from airflow.operators.python import PythonOperator, ShortCircuitOperator | |
| from airflow.utils.trigger_rule import TriggerRule | |
| from airflow.exceptions import AirflowSkipException | |
| from airflow.models import Variable | |
| from airflow.utils.task_group import TaskGroup | |
| import os | |
| from airflow.exceptions import AirflowSkipException | |
| # Utilies | |
| def getBasename(path): | |
| """ | |
| Input: "a/b/c.d.txt" | |
| Output: ("a/b/c.d", ".txt") | |
| """ | |
| return os.path.splitext(path)[0] | |
| def nameToKey(path): | |
| return path.replace("/", "_").replace(".", "_") | |
| def fileExist(**context): | |
| # https://stackoverflow.com/questions/51725746/airflow-run-a-task-when-some-upstream-is-skipped-by-shortcircuit | |
| print(context['params']) | |
| if os.path.exists(context['params'].get('file')): | |
| raise AirflowSkipException(f"Skip.") | |
| return True | |
| def fileExistOperator(file_path): | |
| return ShortCircuitOperator( | |
| task_id='check_exist_' + nameToKey(file_path), | |
| python_callable=fileExist, | |
| params={ | |
| 'file': file_path | |
| }, | |
| trigger_rule=TriggerRule.NONE_FAILED, | |
| ) | |
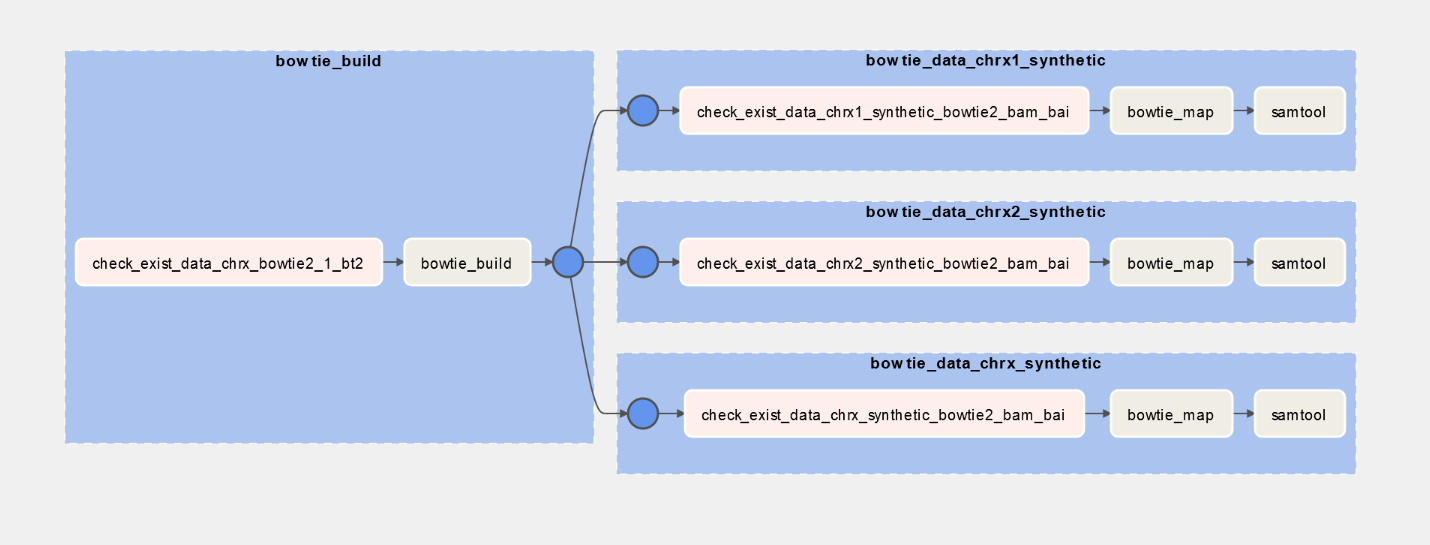
| # Bowtie mapping DAG | |
| def bowtieMap(name): | |
| key = name.replace("/", "_").replace(".", "_") | |
| with TaskGroup(group_id=f'bowtie_{key}') as tg: | |
| map_exist = fileExistOperator(name + ".bowtie2.bam.bai") | |
| templated_command = dedent("""\ | |
| podman run -it --rm -v {{ var.value.basic_folder }}:/app -w /app {{ params.image }} \ | |
| bowtie2 --threads 32 -x {{ var.value.reference_fa | getBasename }}.bowtie2 \ | |
| """) | |
| templated_command += dedent(f"""\ | |
| -1 { name }.read1.fq -2 { name }.read2.fq \ | |
| -S { name }.bowtie2.sam | |
| """) | |
| task_map = BashOperator( | |
| task_id=f'bowtie_map', | |
| bash_command=templated_command, | |
| params={ | |
| 'image': 'quay.io/biocontainers/bowtie2:2.4.4--py39hbb4e92a_0', | |
| }, | |
| ) | |
| map_exist >> task_map | |
| name = name + ".bowtie2" | |
| templated_command = dedent("""\ | |
| podman run -it --rm -v {{ var.value.basic_folder }}:/app -w /app {{ params.image }} \ | |
| """) | |
| templated_command += dedent(f"""\ | |
| samtools sort -@32 {name}.sam -o {name}.bam | |
| """) | |
| templated_command += dedent("""\ | |
| podman run -it --rm -v {{ var.value.basic_folder }}:/app -w /app {{ params.image }} \ | |
| """) | |
| templated_command += dedent(f"""\ | |
| samtools index -@32 {name}.bam | |
| """) | |
| task_samtool = BashOperator( | |
| task_id=f'samtool', | |
| bash_command=templated_command, | |
| params={ | |
| 'image': "quay.io/biocontainers/samtools:1.10--h9402c20_2", | |
| }, | |
| ) | |
| task_map >> task_samtool | |
| return tg | |
| # Bowtie index DAG | |
| def bowtieBuild(reference_fa): | |
| with TaskGroup(group_id=f'bowtie_build') as tg: | |
| index_exist = fileExistOperator(getBasename(reference_fa) + ".bowtie2.1.bt2") | |
| templated_command = dedent(""" | |
| podman run -it --rm -v {{ var.value.basic_folder }}:/app -w /app {{ params.image }} \ | |
| bowtie2-build --threads 32 {{ var.value.reference_fa }} {{ var.value.reference_fa | getBasename }}.bowtie2 | |
| """) | |
| task_build = BashOperator( | |
| task_id='bowtie_build', | |
| bash_command=templated_command, | |
| params={ | |
| 'image': 'quay.io/biocontainers/bowtie2:2.4.4--py39hbb4e92a_0', | |
| }, | |
| ) | |
| index_exist >> task_build | |
| return tg | |
| # main | |
| with DAG( | |
| 'bowtie_all', | |
| description='Build index and read mapping', | |
| start_date=datetime(2021, 1, 1), | |
| schedule_interval=None, | |
| user_defined_filters={ | |
| 'getBasename': getBasename | |
| } | |
| ) as dag: | |
| dag_build = bowtieBuild(Variable.get("reference_fa")) | |
| dag_maps = [] | |
| # Read variable to build DAG, it's Dynamic !! | |
| for name in Variable.get("reads").split(','): | |
| dag_maps.append(bowtieMap(name)) | |
| dag_build >> dag_maps |
Author
linnil1
commented
Jan 10, 2022
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment