A few days ago I used GitHub Actions on a project of mine for the first time. Overall the outcome was very positive, but there was also some work involved. Hopefully, this post can help you do the same at a lower cost!
Create a .github/workflows directory at the root of your project:
mkdir -p .github/workflowsand, in that directory, create one or more YAML file with the name of your choice.
In my case I created two files:
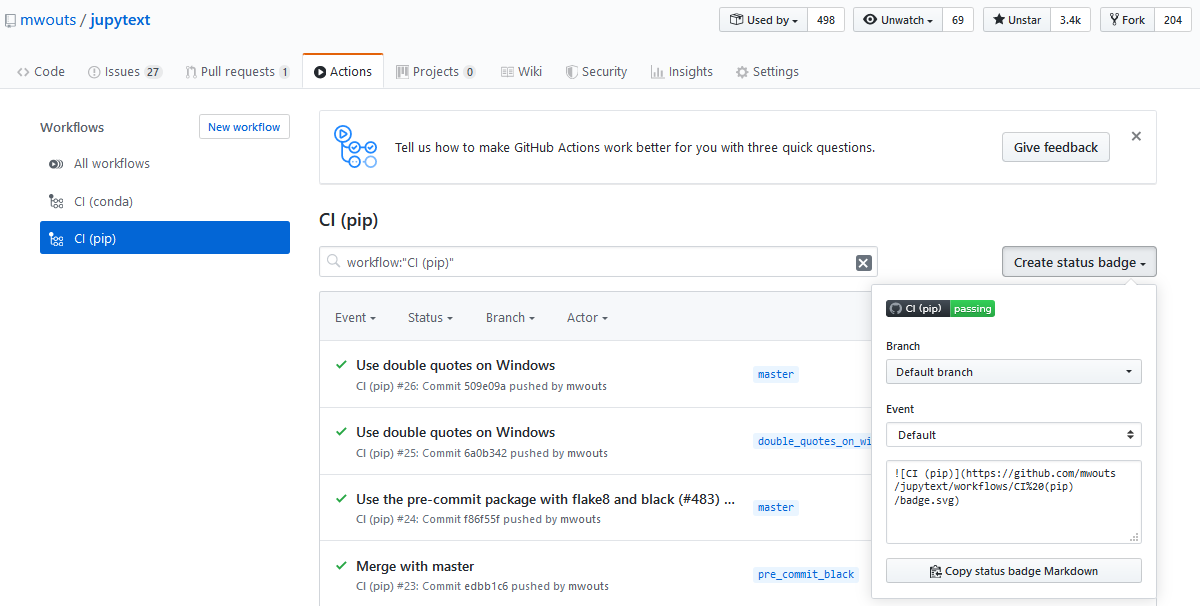
At the top of the YAML file, we have this field: name: CI (pip). While the name of the YAML file does not seem to have any impact, the name field at the top of the file is the one that appears in your Actions tab, and also the name that you will see on your badge:
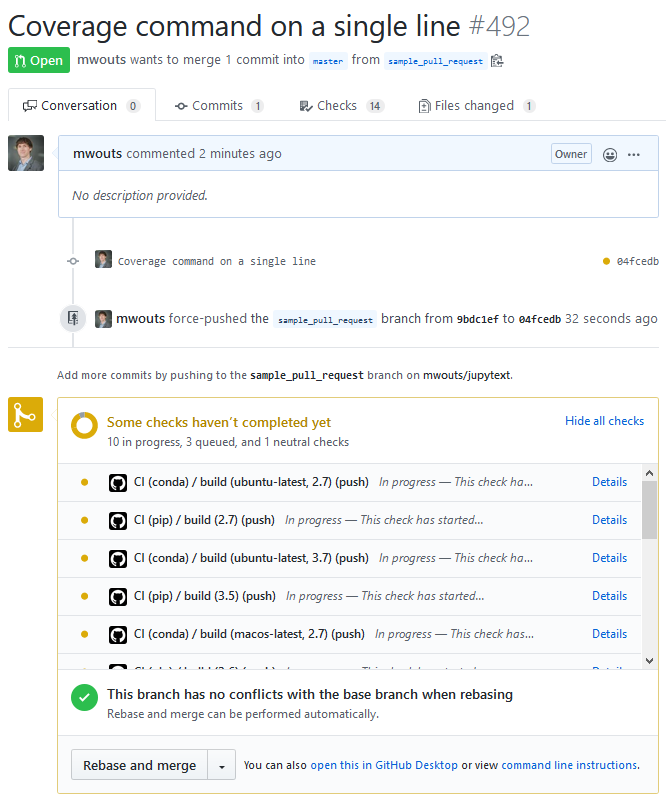
With the on field, you choose which events should trigger an action. I initially tried on: [push, pull_request], but I soon had the feeling that too many builds were triggered. Now I use on: [push] and that seems to be enough. With this, contributors receive an email when one of their commits breaks the CI, so they can fix the issue before opening a pull request. And with on: [push] already, you get status updates in the pull request:
And, a few minutes later:
Python code is portable, but in practice, I prefer to test my code on various platforms.
In my project, I want to test
- Python environments build with both
pipandconda - Python versions 2.7, 3.5, 3.6, 3.7 and 3.8
- Python on Linux, Mac OS or Windows.
Note that I don't need to test the full matrix (30 jobs). If I test all the Python versions with pip and Linux, and Python 2.7 and 3.7 with conda on all the possible OS (11 jobs), then I can already be confident that my program is going to work almost everywhere.
Let's start with pip, which is super easy to set up. Here is an extract of my continuous-integration-pip.yml file, that shows:
- how to check out the GitHub repository with
actions/checkout@v2 - how to install Python with
actions/setup-python@v1, in a version parametrized bypython-version: ${{ matrix.python-version }} - how to install packages from the project's
requirements.txtandrequirements-dev.txtfiles - how to install optional dependencies
name: CI (pip)
on: [push]
jobs:
build:
strategy:
matrix:
python-version: [2.7, 3.5, 3.6, 3.7, 3.8]
runs-on: ubuntu-latest
steps:
- name: Checkout
uses: actions/checkout@v2
- name: Set up Python ${{ matrix.python-version }}
uses: actions/setup-python@v1
with:
python-version: ${{ matrix.python-version }}
- name: Install dependencies
run: |
python -m pip install --upgrade pip
pip install -r requirements.txt
pip install -r requirements-dev.txt
# install black if available (Python 3.6 and above)
pip install black || true
Now we head on to conda. This extract of my continuous-integration-conda.yml file shows:
- how to check out your GitHub repository with
actions/checkout@v2 - how to choose the OS among Ubuntu (Linux), Mac OS, Windows
- how to install Miniconda with
goanpeca/setup-miniconda@v1, with a version of Python parametrized bypython-version: ${{ matrix.python-version }} - how to create the conda environment from the
environment.ymlfile - and how to activate the corresponding environment
name: CI (conda)
on: [push]
jobs:
build:
strategy:
matrix:
os: ['ubuntu-latest', 'macos-latest', 'windows-latest']
python-version: [2.7, 3.7]
runs-on: ${{ matrix.os }}
steps:
- name: Checkout
uses: actions/checkout@v2
- name: Setup Miniconda
uses: goanpeca/setup-miniconda@v1
with:
auto-update-conda: true
auto-activate-base: false
miniconda-version: 'latest'
python-version: ${{ matrix.python-version }}
environment-file: environment.yml
activate-environment: jupytext-dev
Before running any test, it is a good idea to make sure that all the code is valid, which I do with flake8.
The corresponding step in my continuous-integration-pip.yml file is
- name: Lint with flake8
run: |
# stop the build if there are Python syntax errors or undefined names
flake8 . --count --select=E9,F63,F7,F82 --show-source --statistics
# all Python files should follow PEP8 (except some notebooks, see setup.cfg)
flake8 jupytext tests
# exit-zero treats all errors as warnings. The GitHub editor is 127 chars wide
flake8 . --count --exit-zero --max-complexity=10 --statistics
In the continuous-integration-conda.yml, an additional shell parameter was required. I first tried shell: pwsh (PowerShell), and it worked, so I am now using
- name: Lint with flake8
shell: pwsh
run: |
# stop the build if there are Python syntax errors or undefined names
flake8 . --count --select=E9,F63,F7,F82 --show-source --statistics
# all Python files should follow PEP8 (except some notebooks, see setup.cfg)
flake8 jupytext tests
# exit-zero treats all errors as warnings. The GitHub editor is 127 chars wide
flake8 . --count --exit-zero --max-complexity=10 --statistics
I had initially an issue with the flake8 step. The error was:
Lint with flake8
4s
##[error]Process completed with exit code 2.
Run # stop the build if there are Python syntax errors or undefined names
# stop the build if there are Python syntax errors or undefined names
flake8 . --count --select=E9,F63,F7,F82 --show-source --statistics
all Python files should follow PEP8 (except some notebooks, see setup.cfg)
flake8 jupytext tests
# exit-zero treats all errors as warnings. The GitHub editor is 127 chars wide
flake8 . --count --exit-zero --max-complexity=10 --statistics
shell: /bin/bash -e {0}
env:
pythonLocation: /opt/hostedtoolcache/Python/3.6.10/x64
0
/home/runner/work/_temp/98d1db20-f0af-4eba-af95-cb39421c77b0.sh: line 3: syntax error near unexpected token `('
##[error]Process completed with exit code 2.
My understanding is that /home/runner/work/_temp/98d1db20-f0af-4eba-af95-cb39421c77b0.sh is a temporary script that contains the commands for that step.
So when we are told that the script throws this error: line 3: syntax error near unexpected token `(', we should look at the third line of the run
property of that step. In my command, that was all Python files should follow PEP8 (except some notebooks, see setup.cfg), and indeed it was missing a
comment char!
Once we are positive that the code is syntactically correct, we want to know whether all the unit tests pass. The corresponding step is:
- name: Test with pytest
run: coverage run --source=. -m py.test
Note that I use coverage run --source=. -m py.test and not just pytest because I also want to know the code coverage. Also, for the conda file, we need to add a shell: pwsh property, otherwise, the coverage or pytest command is not found.
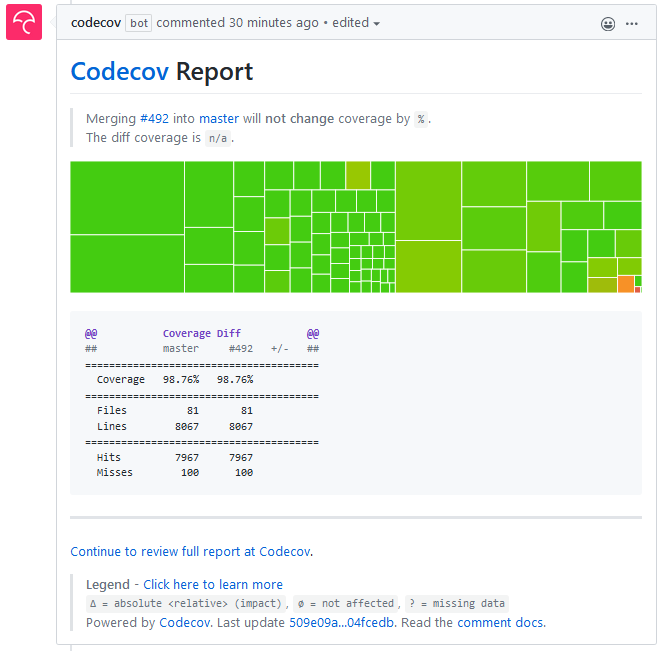
It's good to have the coverage computed in the CI, but it is even better to have a coverage update on pull requests, and a coverage badge in the README. For this, I use codecov. I prefer to upload the coverage only for the conda CI as it lets me test a larger fraction of the optional features. The coverage upload step is the last one in continuous-integration-conda.yml:
- name: Upload coverage
shell: pwsh
run: coverage report -m
With this, I get a coverage badge which I can add to my README, detailed coverage statistics and charts at codecov.io, as well as coverage reports in pull requests:
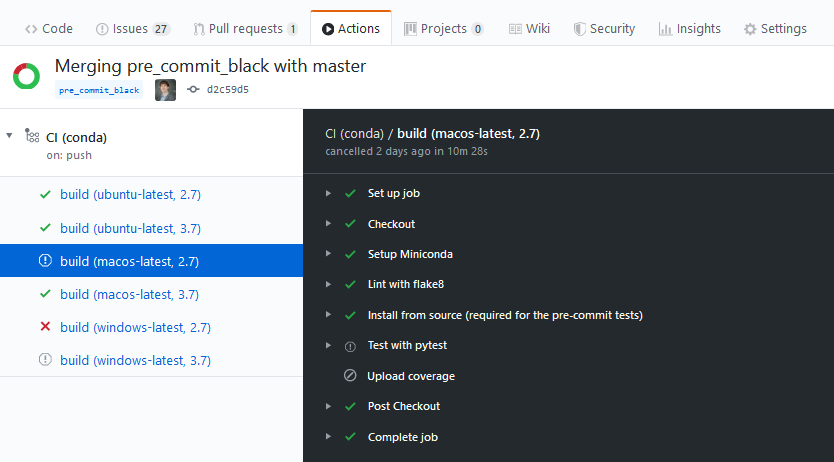
One feature that surprised me at first, but makes sense, is the automated job cancellation. When one job in the CI fails, all the other jobs that are still running or pending are canceled.
Here is an example of this on my project - a problem occurs with the Windows build, and that triggers the cancellation of the remaining Windows and Mac OS jobs:
Before discovering GitHub Actions, I was using Travis-CI. And I loved it! I have used Travis-CI on many projects, including this fancy one where we test... the README itself!
Now I will conclude with a brief comparison between the two.
- Travis-CI has been there for a much longer period. I could have feared that documentation would be harder to find for GitHub Actions, but that was not the case. Useful references were: Using Python with GitHub Actions and Setup Miniconda.
- Just like Travis-CI, Github Actions is free to use for public projects.
- GitHub integration is excellent for both Travis-CI and GitHub Actions, although the Actions have a dedicated tab on the project page, which makes it even easier to access the CI.
- Configuring conda with Github Actions turned out to be simpler than with Travis-CI, for which I was using this hack.
- I found the Matrix (e.g. Python version times OS like here) simpler to use on Github Actions.
- I have not noticed a significant difference in the duration of the jobs. On my sample PR, the six jobs (pip and conda, Linux only) on Travis-CI ran in 8m 33s, while on GitHub Actions, the first series of five pip jobs (Linux only) ran in 4m 57s, while the other series of six conda jobs (Linux, Mac OS, Windows) ran in parallel in 12m 48s - so, in that example, GitHub Actions took 50% longer, but also covered a larger number of configurations (+80%).
Thanks for reading this! If you want to read more, have a look at my other posts on Medium, at my open source projects on GitHub, or follow me on Twitter. Also, if you have experience with GitHub Actions in other contexts, if you know e.g. how to publish documentation online, please let us know with a comment here, or feel free to open an issue or make a pull request on this repo!