Last active
June 10, 2018 15:06
-
-
Save quantum-kite/3218dab366b30bbe3040726cfecdd018 to your computer and use it in GitHub Desktop.
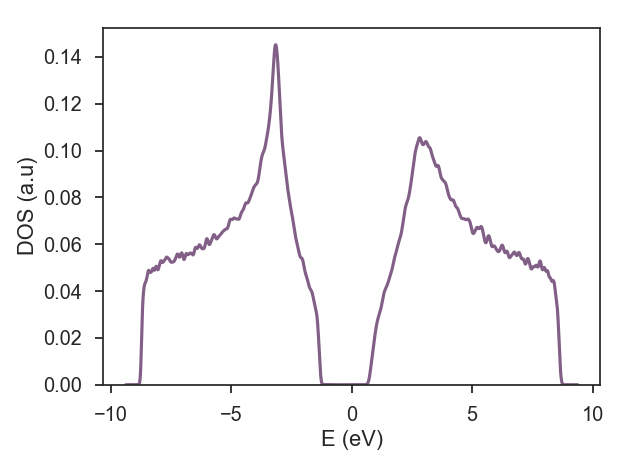
example of adding on-site disorder
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| """ Onsite disorder | |
| Lattice : Monolayer graphene; | |
| Disorder : Disorder class Deterministic and Uniform at different sublattices, | |
| Configuration : size of the system 512x512, without domain decomposition (nx=ny=1), periodic boundary conditions, | |
| double precision, manual scaling; | |
| Calculation : dos; | |
| Modification : magnetic field is off; | |
| """ | |
| import kite | |
| from pybinding.repository import graphene | |
| # load graphene lattice and structural_disorder | |
| lattice = graphene.monolayer() | |
| # add Disorder | |
| disorder = kite.Disorder(lattice) | |
| disorder.add_disorder('B', 'Deterministic', -1.0) | |
| disorder.add_disorder('A', 'Uniform', +1.5, 1.0) | |
| # number of decomposition parts in each direction of matrix. | |
| # This divides the lattice into various sections, each of which is calculated in parallel | |
| nx = ny = 1 | |
| # number of unit cells in each direction. | |
| lx = ly = 512 | |
| # make config object which caries info about | |
| # - the number of decomposition parts [nx, ny], | |
| # - lengths of structure [lx, ly] | |
| # - boundary conditions, setting True as periodic boundary conditions, and False elsewise, | |
| # - info if the exported hopping and onsite data should be complex, | |
| # - info of the precision of the exported hopping and onsite data, 0 - float, 1 - double, and 2 - long double. | |
| # - scaling, if None it's automatic, if present select spectrum_bound=[e_min, e_max] | |
| configuration = kite.Configuration(divisions=[nx, ny], length=[lx, ly], boundaries=[True, True], | |
| is_complex=False, precision=1) | |
| # require the calculation of DOS | |
| calculation = kite.Calculation(configuration) | |
| calculation.dos(num_points=5000, num_moments=512, num_random=1, num_disorder=1) | |
| # configure the *.h5 file | |
| kite.config_system(lattice, configuration, calculation, modification, 'on_site_disorder.h5', disorder=disorder) |
Author
quantum-kite
commented
Jun 5, 2018
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment