Last active
July 17, 2018 10:26
-
-
Save samronsin/7177d1061f1030166dde754322b39e6c to your computer and use it in GitHub Desktop.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| import numpy as np | |
| from sklearn.metrics import calibration_loss | |
| def sample_calibrated(N): | |
| xs = [] | |
| ys = [] | |
| for _ in range(N): | |
| x = np.random.rand() | |
| xs.append(x) | |
| y = 1 if np.random.rand() < x else 0 | |
| ys.append(y) | |
| x_arr = np.array(xs) | |
| y_arr = np.array(ys) | |
| return y_arr, x_arr | |
| # Compare calB (overlapping) and loss based on non-overlapping bins | |
| ratio = 0.1 | |
| calBs = [] | |
| calCs = [] | |
| ks = range(2, 7) | |
| for k in ks: | |
| N = 10 ** k | |
| y_arr, x_arr = sample_calibrated(N) | |
| calB = calibration_loss(y_arr, x_arr, bin_size_ratio=ratio, sliding_window=True) | |
| calBs.append(calB) | |
| calC = calibration_loss(y_arr, x_arr, bin_size_ratio=ratio, sliding_window=False) | |
| calCs.append(calC) | |
| plt.plot(ks, calBs) | |
| plt.plot(ks, calCs, c="r") | |
| # Compare different values of bin_size_ratio | |
| cal1s = [] | |
| cal2s = [] | |
| cal3s = [] | |
| ks = range(2, 7) | |
| for k in ks: | |
| N = 10 ** k | |
| y_arr, x_arr = sample_calibrated(N) | |
| cal1 = calibration_loss(y_arr, x_arr, bin_size_ratio=0.1, sliding_window=False) | |
| cal1s.append(cal1) | |
| cal2 = calibration_loss(y_arr, x_arr, bin_size_ratio=0.01, sliding_window=False) | |
| cal2s.append(cal2) | |
| cal3 = calibration_loss(y_arr, x_arr, bin_size_ratio=0.001, sliding_window=False) | |
| cal3s.append(cal3) | |
| plt.plot(ks, cal1s, c="r") | |
| plt.plot(ks, cal2s, c="g") | |
| plt.plot(ks, cal3s, c="b") | |
| # Compare Brier score and calibration loss | |
| from sklearn.metrics import brier_score_loss | |
| ratio = 0.1 | |
| calBs = [] | |
| briers = [] | |
| ks = range(2, 7) | |
| for k in ks: | |
| N = 10 ** k | |
| y_arr, x_arr = sample_calibrated(N) | |
| calB = calibration_loss(y_arr, x_arr, bin_size_ratio=ratio) | |
| calBs.append(calB) | |
| brier = brier_score_loss(y_arr, x_arr) | |
| briers.append(brier) | |
| plt.plot(ks, calBs, c="r") | |
| plt.plot(ks, briers, c="b") |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| import numpy as np | |
| from sklearn.metrics import calibration_loss | |
| def twist(x, e): | |
| if x < 0.5: | |
| return np.power(x, e) / np.power(0.5, e-1) | |
| else: | |
| return 1 - np.power(1-x, e) / np.power(0.5, e-1) | |
| def sample_poorly_calibrated(N, e): | |
| xs = [] | |
| ys = [] | |
| for _ in range(N): | |
| x = np.random.rand() | |
| xs.append(x) | |
| h = twist(x, e) | |
| y = 1 if np.random.rand() < h else 0 | |
| ys.append(y) | |
| x_arr = np.array(xs) | |
| y_arr = np.array(ys) | |
| return y_arr, x_arr | |
| # Plot calibration curve | |
| y_arr, x_arr = sample_poorly_calibrated(1000000, e) | |
| ys, xs = calibration_curve(y_arr, x_arr, n_bins=100) | |
| plt.plot(xs, ys) | |
| # Compare calB (overlapping) and loss based on non-overlapping bins | |
| ratio = 0.1 | |
| e = 8 | |
| calBs = [] | |
| calCs = [] | |
| ks = range(2, 7) | |
| for k in ks: | |
| N = 10 ** k | |
| y_arr, x_arr = sample_poorly_calibrated(N, e) | |
| calB = calibration_loss(y_arr, x_arr, bin_size_ratio=ratio, sliding_window=True) | |
| calBs.append(calB) | |
| calC = calibration_loss(y_arr, x_arr, bin_size_ratio=ratio, sliding_window=False) | |
| calCs.append(calC) | |
| plt.plot(ks, calBs, c="b") | |
| plt.plot(ks, calCs, c="r") |
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment

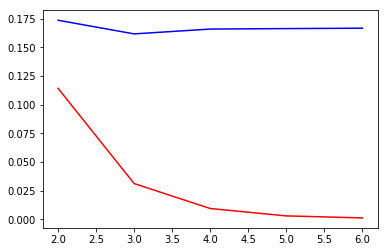
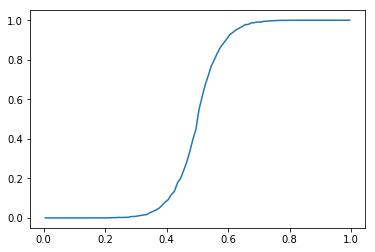
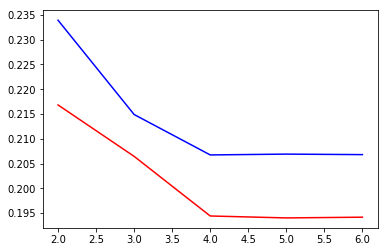
sliding_windowdoes not make much of a difference...