Table of Contents
- Introduction
- Optimizing performance in Python
- What does "Optimizing code" mean anyway?
- When to optimize?
- Identifying performance bottlenecks
- Profilers in Python
- Profiling CPU performance (and execution time) in Python
- How do CPU profilers work in Python?
- Approach for optimizing performance
- Some caveats (must know things) about profilers and optimizations
- Where and how an APM (Application Performance Monitoring) tool help here?
- Conclusion
Title: Identifying performance bottlenecks in your Python code and optimizing it
In this post, we will walk through various techniques that can be used to identify the performance bottlenecks in your python codebase and optimize them.
The term "optimization" can apply to a broad level of metrics. But two general metrics of most interest are; CPU performance (execution time) and memory footprint. For this post, you can think of an optimized code as the one which is either able to run faster or use lesser memory or both.
There are no hard and fast rules. But generally speaking, you should try optimizing when you are sure that the business logic in the code is correct and not going to change soon.
"First make it work. Then make it right. Then make it fast." ~ Kent Beck
Otherwise, the effort spent on premature-optimization can go futile.
Unless you're developing a performance-intensive product or a code dependency that is going to be used by other projects which might be performance-intensive, optimizing every aspect of the code can be an overkill. For most of the scenarios, the 80-20 principle (80 percent of performance benefits may come from optimizing 20 percent of your code) will be more appropriate. According to professor Donald Knuth,
Programmers waste enormous amounts of time thinking about or worrying about, the speed of noncritical parts of their programs, and these attempts at efficiency actually have a strong negative impact when debugging and maintenance are considered. We should forget about small efficiencies, say about 97% of the time: premature optimization is the root of all evil. Yet we should not pass up our opportunities in that critical 3%.
Before writing the optimized version of the code, we need to know what to optimize, "What is that 3% I should be spending my effort on?", or in other words, we need to identify the bottlenecks,
Most of the times we make intuitive-guesses, but more often than not, our guesses are either wrong or just approximately correct. So, it's always advisable to take help of tools to get a clear definitive picture. The process of measuring resources (CPU usage, RAM, I/O, etc.) used by different sections of a program is known as "profiling," and the tools that help in profiling are usually referred to as "profilers." A profiler typically collects statistics like how often a resource is used and who is using the resource. For instance, a profiler designed for profiling execution time will measure how often and for how various long parts of the code are executed. Using a profiling mechanism becomes a necessity when the codebase grows large, and you still want to maintain efficiency.
The world of profilers in Python is pretty large, and if it's the first time that you decided to profile something, you may get overwhelmed by the number of options that you'll have. But the nice thing is some profilers are best suited for particular cases, understanding this can help you avoid a lot of stress when deciding which profiler to use, and this is exactly what we're going to cover in next few sections.
There are multiple criteria based on which you can segregate profilers in python,
A profiler collects data and statistics by monitoring the events (function calls, returns, exceptions, etc.) that happen during program execution. There are broadly two approaches to go about it, either the profiler will be monitoring all the events, or it will sample after time intervals to collect that information. The former one is called deterministic profiling, and the later one is called statistical profiling. The tradeoff that we are making in these two approaches is between overhead and accuracy. You may find the terms deterministic and statistical profilers used interchangeably with "tracing" and "sampling" profilers, but they're not the same technically. Tracing simply means recording the entire call graph, and sampling simply means probing in intervals.
Some profilers will measure resource usage at the module level, some are capable of doing it at a function level, and some can measure at line level.
There are some tools which aren't profilers themselves, but they provide an interface to perform analysis on the data collected / reports generated by other popular profilers.
This section will walk you though various options that are available to us. Wherever possible, we'll group similar profilers for simplification.
The timing modules (time module and the timeit) are the basic modules that you can use to measure the execution time of various approaches (kind of algorithmic testing). The usage is pretty simple.
import time
start_time = time.time()
# The code to be evaluated
end_time = time.time()
# Time taken in seconds
time_taken = end_time - start_timeHowever, there are a few external factors that can affect time_taken. To minimize the influence of those external factors (e.g., machine load, I/O), it is often advised to run the test multiple times and choose the fastest.
Another thing to note is that time.time() measures the wall-clock time ( total time taken for the code to get completely executed) and not the CPU time (total time for which the CPU was actually processing the statements related to the code, excluding the time where it was busy somewhere else). To measure CPU time, you can use time.clock() instead.
timeit module was designed to abstract a lot of such intricacies and provide a simple interface for measuring execution times. Here's a sample usage
>>> import timeit
>>> timeit.timeit(stmt='" ".join(some_list)', setup='some_list = ["lets", "join", "some", "strings"]', number=1000, timer=time.clock)
0.00020100000000011775The above code will run the setup statement once, and then returns the cumulative wall-clock time taken to run the main statement (stmt) 1000 times.
You can also use timeit.repeat to run the same experiment multiple time. And to measure CPU time instead of wall clock time, you can pass the timer as time.process_time). Here's an example,
>>> timeit.repeat(stmt='" ".join(some_list)', setup='some_list = ["lets", "join", "some", "strings"]', number=1000, repeat=4, timer=time.process_time)
[0.00019499999999972317,
0.0001930000000003318,
0.00019100000000005224,
0.00019799999999969842]Also, if you're using jupyter notebook, you can use the %timeit magic function to get similar (and nicely formatted) results.
The built-in profile and cProfile modules provide deterministic profiling. Both have a similar interface, but profile is implemented in pure python, which adds to the overhead.
Usage:
$ python -m profile somefile.py *args
$ python -m cProfile somefile.py *argsWhere *args are the command line arguments to be passed to your python file (if any).
This will print out results on stdout in the following format.
844321 function calls (711197 primitive calls) in 7.343 seconds
Ordered by: standard name
ncalls tottime percall cumtime percall filename:lineno(function)
992 0.001 0.000 0.001 0.000 json_generator.py:49(is_interactive_statement)
You can also use the -o option to save to the results to file so that we can use them later.
What do the different columns in results mean?
ncalls is the number of calls, tottime is the time spent in a given function excluding the time spent in calls to sub-functions, percall is the time per call for a function (essentially tottime divided by ncalls), cumtime is the cumulative time spent in the function including the time spent in calls to sub-functions, the percall next to it is again the per call cumulative time, and finally, filename:lineno provides the reference to the code (at function level) related to these stats.
This and this were the sample outputs when I tried to run them on a couple of my scripts. As you can observe, there is plenty of information to go through in those outputs. If you're interested in only a subset of information, you can also use them as a python module to profile specific portions of the code.
import cProfile
p = cProfile.Profile()
# Enable profiling
p.enbale()
# Some logic that you want to profile
# Disable profiling
p.disable()
# Print the stats
p.print_stats()
# Dump the stats to a file
p.dump_stats("results.prof")The interface of profile module is slightly different, and it also has a calibration method for correcting the overhead time from results. If you are not extending the profiler and just want to see the stats, you should always use cProfile instead of profile as it has less overhead.
However, there are few cases where cProfile and profile modules won't be helpful for you,
- Event loop based applications; cProfile doesn't understand gevent or any greenlet based application.
- Line by line profiling; Sometimes you will want to know the lines from which your time-consuming function is being called, and sometimes you may have difficulty in figuring out which line inside the time-consuming-function is the slowest.
- Lack of visualization features. No support of exporting the stats to formats like callgrind.
- Tricky do certain things like measuring CPU time, profiling multithreading programs, and so on. By default, the CProfiler runs on the main thread only (thread on which your python script is invoked). If you want to profile other threads that are spawned from your program, you might want to use
threading.setprofilemethod of the threading module.
Nothing to worry here though, there are other tools that can complement cProfile or serve as an alternative to cProfiler in some cases. Let's discuss them next,
The pstats module can be used to parse the data generated by profile or cProfile modules. Once parsed you can do things like
- Sorting based on number of calls, time, cumulative time, etc
- Combining stats from multiple sources
- Stripping directory names from the data
- Viewing limited data (based on criterion like top n, containing regex, etc.)
- Finding callers and callees of a function
Here's the typical usage
import pstats
# parse the results of cProfile
s = pstats.Stats("results.prof")
# Adding stats
s.add("more_results.prof")
# remove directory paths
s.strip_dirs()
# Sorting based on cumulative time
s.sort_stats("cumulative")
# Printing top 'n' stats
n = 3
s.print_stats(n)
function_name = "foo"
# print callers of foo
stats.print_callers('\({}'.format(function_name))
# pring callees of foo
stats.print_callers('\({}'.format(function_name))line_profiler performs line by line profling collecting stats about the time spent in each line of the code being profiled.
Usage
$ pip install line_profiler# You python program
# Use the decorator
@profile
def some_func(*args, **kwargs)
...# kernprof is a convenient script to run different profilers; it comes with line_profiler
# https://github.com/rkern/line_profiler/blob/master/kernprof.py
$ kernprof -l -v python_script.py
# This will generate a binary file named python_script.py.lprof
# You can visaulize this file by
$ python -m line_profiler python_script.py.lprofHere's a sample output (complete output here)
Timer unit: 1e-06 s
Total time: 0.000825 s
File: json_generator.py
Function: generate_code_block at line 26
Line # Hits Time Per Hit % Time Line Contents
==============================================================
26 @profile
27 def generate_code_block(statements, output):
28 global sequence_num
29 result = {
30 278 109.0 0.4 13.2 "type": "code",
31 278 143.0 0.5 17.3 "sequence_num": sequence_num,
32 278 109.0 0.4 13.2 "statements": statements,
33 278 220.0 0.8 26.7 "output": output
34 }
35 278 130.0 0.5 15.8 sequence_num += 1
36 278 114.0 0.4 13.8 return result
Total time: 0.014138 s
File: json_generator.py
Function: parse_example_parts at line 55
Line # Hits Time Per Hit % Time Line Contents
==============================================================
55 @profile
56 def parse_example_parts(lines, example_title_line):
57 parts = {
58 57 38.0 0.7 0.3 "build_up": [],
The column names are kind of self-explanatory. Hits are the number of times the line is executed during the run, and "% time" is the propotion of time spent on the line. (It'll sum up to 100 for every function).
Yappi (Yet Another Python Profiler) is a profiler which is an attempt to improve upon some of the lacking features of cProfile. Yappi can,
- Profile a multi-threaded python application out of the box. It also provides the functionality of obtaining per-thread function statistics.
- Measure CPU time easily.
- Export the results to callgrind format as well as pstat formats.
Usage
$ pip install yappiimport yappi
# The default clock is set to CPU, but you can switch to wall clock
# yappi.set_clock_type("wall")
yappi.start()
# Call your functions or perform execution here
some_func()
# Getting all the stats
yappi.get_func_stats().print_all()Here's the sample output (complete output here).
name ncall tsub ttot tavg
../lib/python3.6/pprint.py:47 pprint 2 0.000050 5.181650 2.590825
..pprint.py:138 PrettyPrinter.pprint 2 0.000043 5.181576 2.590788
..print.py:154 PrettyPrinter._format 101.. 0.234201 5.181516 0.000508
...py:180 PrettyPrinter._pprint_dict 148.. 0.047156 4.244369 0.002868
..3 PrettyPrinter._format_dict_items 148.. 0.149224 4.232629 0.002860
...py:207 PrettyPrinter._pprint_list 999/2 0.016574 4.169317 0.004173
..py:350 PrettyPrinter._format_items 999/2 0.086727 4.169248 0.004173
..nerator.py:226 convert_to_notebook 1 0.001365 3.705809 3.705809
../pprint.py:391 PrettyPrinter._repr 15608 0.192905 3.533774 0.000226
..pprint.py:400 PrettyPrinter.format 15608 0.120687 3.277539 0.000210
..python3.6/pprint.py:490 _safe_repr 931.. 1.554210 3.156852 0.000034
..ython3.6/json/__init__.py:120 dump 1 0.124881 0.937511 0.937511
...6/json/encoder.py:412 _iterencode 19093 0.116629 0.749296 0.000039
..on/encoder.py:333 _iterencode_dict 488.. 0.329734 0.632647 0.000013
..t.py:244 PrettyPrinter._pprint_str 1020 0.277267 0.576017 0.000565
..python3.6/pprint.py:94 _safe_tuple 30235 0.352826 0.559835 0.000019
..on/encoder.py:277 _iterencode_list 297.. 0.201149 0.515704 0.000017
...6/pprint.py:84 _safe_key.__init__ 60470 0.207009 0.207009 0.000003
..n3.6/pprint.py:87 _safe_key.__lt__ 32160 0.114377 0.114377 0.000004
..enerator.py:60 parse_example_parts 57 0.043338 0.104288 0.001830
.._generator.py:173 convert_to_cells 112 0.004576 0.044347 0.000396
..62 inspect_and_sanitize_code_lines 278 0.011167 0.037915 0.000136Just like cProfiler, the metrics are collected at function level. tsub is the total time spent in the function excluding the subcalls, ttot is the total time including them, ncall is the number of calls, tavg is the per call time (ttot divided by ncall).
hotshot is also a built-in module for profiling. The difference between hotshot and cProfile (or profile) is that hotshot doesn't interfere as much as cProfile does. The idea behind hotshot is to minimize the profiling overhead by doing all the data processing at the end. However, the module is no longer maintained, and the python docs say that it might be dropped in future versions.
The interface of hostshot is similar to that of cProfile; you can read the docs here.
coldshot was an attempt to improve upon hotshot; the major feature is the ability to visualize results in a tool like RunSnakeRun (discussed later).
vmprof-python is a statistical line profiler for Python. Due to statistical nature, vmprof-python has a low overhead.
Usage
$ pip install vmprof
$ python -m vmprof --lines -o results.dat somefile.py
$ vmprofshow --lines results.datYou can omit the --lines option to profile at function level rather. Follwing is the sample output (complete output here)
Line # Hits % Hits Line Contents
=======================================
226 def convert_to_notebook(parsed_json):
227 result = {
228 "cells": [],
229 "metadata": {},
230 "nbformat": 4,
231 "nbformat_minor": 2
232 }
233 for example in parsed_json:
234 parts = example["parts"]
235 build_up = parts.get("build_up")
236 explanation = parts.get("explanation")
237 notebook_path = "test.ipynb"
238
239 if(build_up):
240 4 2.4 result["cells"] += convert_to_cells(build_up)
241
242 if(explanation):
243 result["cells"] += convert_to_cells(explanation)
244
245 153 90.0 pprint.pprint(result, indent=2)
246 1 0.6 with open(notebook_path, "w") as f:
247 12 7.1 json.dump(result, f)
Since it's a statistical profiler and collects data by probing the stack frame in fixed intervals, it might not be as accurate as expected particularly in the cases when there are not enough repetitions (or recursion), and the program runs quickly.
pyinstrument is another actively maintained statistical profiler that records the call stack every 1ms. pyinstrument can measure CPU time, and it also supports full-stack recording, meaning, it generates output like (expanded output generated using --show-all option here:
_ ._ __/__ _ _ _ _ _/_ Recorded: 23:00:06 Samples: 583
/_//_/// /_\ / //_// / //_'/ // Duration: 3.023 CPU time: 0.519
/ _/ v3.0.3
Program: json_generator.py
3.022 <module> json_generator.py:11
├─ 1.583 pprint pprint.py:47
│ [137 frames hidden] pprint, re
│ 0.606 _pprint_str pprint.py:244
└─ 1.421 convert_to_notebook json_generator.py:226
├─ 1.381 pprint pprint.py:47
│ [155 frames hidden] pprint, re
└─ 0.034 dump json/__init__.py:120
[17 frames hidden] json
To view this report with different options, run:
pyinstrument --load-prev 2019-06-27T23-00-06 [options]
Usage:
$ pip install pyinstrument
$ python -m pyinstrument some_file.pyIt also provides middlewares for instrumenting Django and flask; you can read more about that here.
Stacksampler is a statistical profiler written in just 100 lines of code. It records stack every 5ms and works well with gevent-based applications. To visualize the results, you can use the complementary stackcollector agent, which will generate the flame graphs for you. A flame graph has the call stack at the y-axis and percent resource utilization at x-axis (CPU time in our case), which enables you to visualize what part of the program is taking what percent of the time. However, due to its simple implementation, there might be some cases (like profiling dynamically spawned processes/threads) where stacksampler won't give the results you want.
Another interesting profiling tool when it comes to generating flame graphs is pyflame (designed by Uber). The functioning of pyflame is different and more optimized from that of stacksampler (discussed later). The coolest feature of the flame graph is the possibility of calculating a "diff" of two flame graphs. This is immensely helpful when your application starts performing slowly in a recent release. By computing the diff, you can see what changed in the call stack, and how much time it is taking, saving time in debugging performance regressions. However, as per the docs,
Are BSD / OS X / macOS Supported? Pyflame uses a few Linux-specific interfaces, so, unfortunately, it is the only platform supported right now.
So I wasn't able to test it out and present the output in this post :(
Werkzeug is a library consisting of various utilities for WSGI web applications. Flask web framework also wraps werkzeug. One utility that werkzeug library provides is Application Profiler middleware. It uses cProfile to profile each request execution. You can see the usage in docs here.
GreenlentProfiler is a profiler suited for profiling gevent based processes. gevent is a coroutine based networking library that provides a high-level synchronous API on top of the libev event loop. It is used to provide concurrency (but no parallelism) to improve speed by executing I/O bound tasks (network calls) asynchronously under the hood. Here's the post by the owner of GreenletProfiler explaining how cProfiler and YAPPI aren't much helpful for gevent based applications (with examples), and to use GreenletProfiler instead.
gprof2dot is a tool that you can use to visualize the results of profilers like cProfiler and hotshot (and lots of other language profilers) into what is called a dot graph.
- Use cProfile / Hotshot to get results in
.dotfile - Has a django fontend for visualizing (called graphi)
Usage
$ pip install gprof2dot
# To install dot on osx
$ brew install graphviz$ python -m cProfile -o results.prof somefile.py
$ gprof2dot -n0 -e0 -f pstats -o results.dot results.prof
$ dot -Tpng -o results.png results.dotHere's the sample output (view full-size image here), which consists of nodes
Here total time % represents the percentage of time spent in the function, self time % is the time spent in the function excluding sub-function calls, and total calls is the total number of calls.
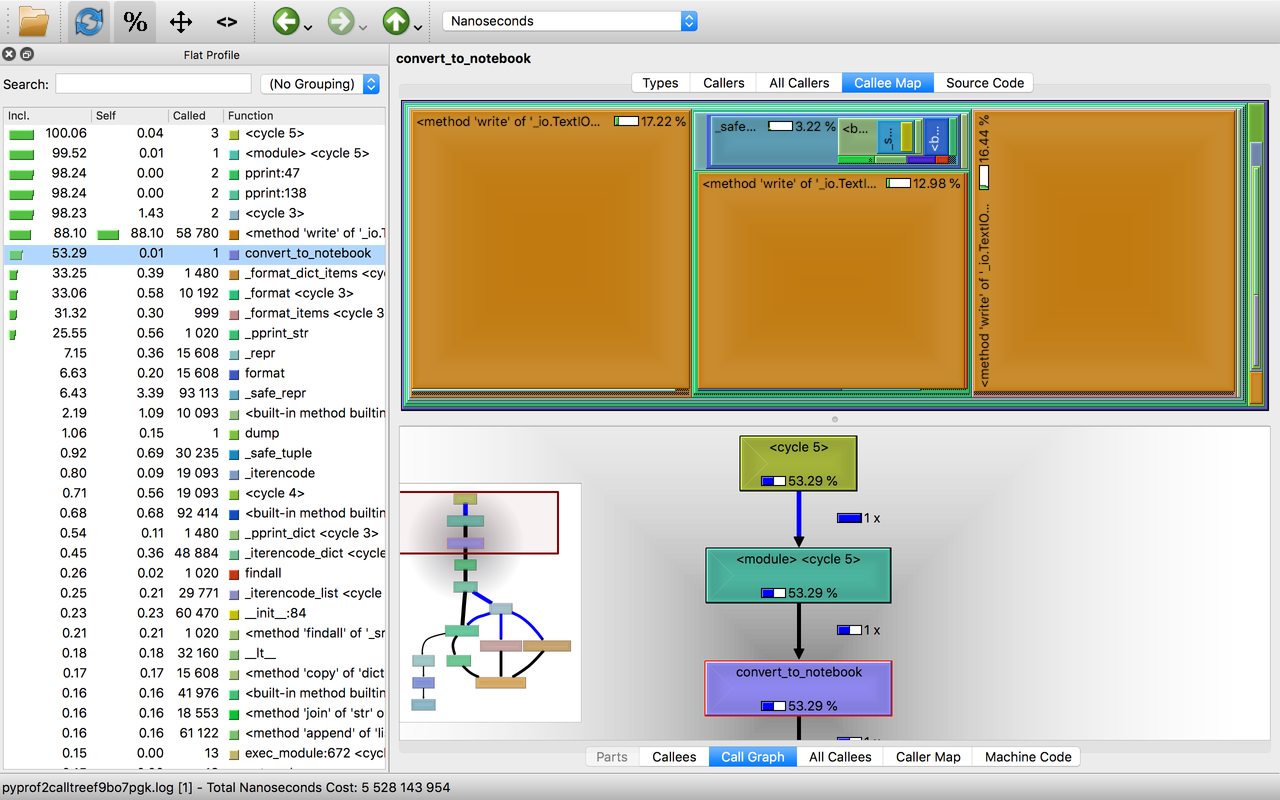
KCacheGrind is a very popular profile data visualization tool. To visualize your python program using it, you can convert cProfile results into the callgrind form using pyprof2calltree and then visualize them using KCacheGrind.
Usage:
$ python -m cProfile -o results.prof somefile.py
$ pyprof2calltree -k -i results.profHere are a couple of screenshots,
You can visualize the call graph, caller graph, callee graph, and even the source code level statistics using kcachegrind.
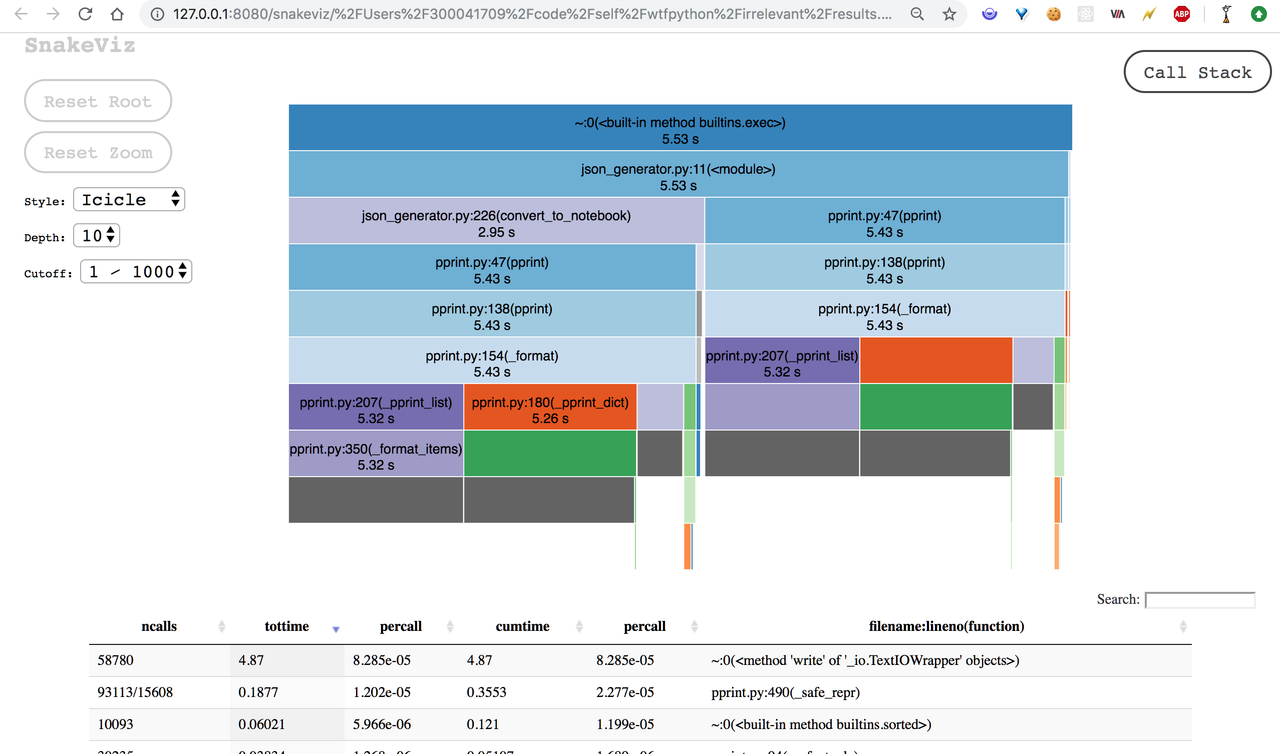
sankeviz is a profile visualizer which lets you convert the output of the cProfile module into browser-based visualizations like sunburst diagrams and nice graphs. Tuna (inspired by SnakeViz) and runsankerun (early tool in this category) are some other options that you can explore.
Snakeviz usage
$ pip install snakeviz
$ python -m cProfile -o results.prof some_file.py
$ snakeviz results.profHere's the screenshot of the output it generates,
Although you don't need to know how the CPU profilers work under the hood to be able to use them, but knowing how they work will help you reason about their capabilities and overheads.
Most of the profilers will function as a part of your Python process and access the stack frames (which will contain meta information like line number, statements, etc.).
The tracing profilers work by registering a callback to interpreter events (function calls, returns, line execution, exceptions, etc.). When the event is triggered, the callback function gets called, which has the logic for recording stack frames and analyzing them. The C-interface functions for registering callbacks are PyEval_SetProfile and PyEval_SetTrace. The difference between the two lies in the events they monitor. Do check out the Profiling and tracing section of the python docs for more details. Here's the ceval.c module in CPython implementation (which by the way, is the heart of python) where these functions are defined.
For checking how they are used, you can see the implementation of the callback functions in the source codes of different tracing profilers. For instance, in the built-in cProfiler, this is the line of code where callback is registered, and this is where the callback function is defined.
Coming to sampling profilers, most of them use setitimer system call in Linux which asks the OS to send signals after fixed intervals. And just like the callback functions in case of tracing profilers, there are signal handlers which will record the stack frame at that moment for analysis. The "fixed intervals" can be wall time or CPU time, depending on the arguments passed to the system call. Here's documentation for settimer. You can explore source codes of different profilers and see how they are using it. For instance, this is the line of code is stacksampler where the settimer signal is registered, this is where the signal handler is defined, and this is the implementation of the signal handler.
Now you can also figure out when and why tracing profilers can have high overhead. If the program that you are profiling generates a lot of events (Example, a recursive function), it will cause the callback function to execute at every event, resulting in overhead. The ovehead of sampling profilers will still be roughly the same in these cases. However, one caveat with OS signal based sampling profilers is that the Signals can interrupt system calls (so that signal handler can run). This can lead to IOErrors and may even conflict with other libraries/program that depends on the same signal the profiler is depending on.
That being said, the differentiation between the implementations of tracing and sampling profilers is not very clear-cut. There are also some sampling profilers like Pyinstrument that have shifted to PyEval_SetProfile approach, wherein the callback function will not record the stack frames until the "fixed interval" has elapsed (to reduce the overhead). And some sampling profilers (like pyflame) run in a different process altogether making use of ptrace system call to record the information for analysis. For those interested, here's a very interseting blog post by Uber engineering on how exactly pyflame works.
Now that we are aware of how profilers work and all the tools that are available to help us in identifying bottlenecks, let's discuss the rough approach that we can follow to eventually optimize our code.
Optimization with the help of profiling is like debugging. A profiler (in most of the cases) won't tell you how to change your code, but it'll help you validate your hypotheses. Usually, the process goes something like this; You identify a culprit section of code, then you come up with some hypothesis
Doing this in X way instead of Y might increase the performance.
And then you try that out to validate your hypotheses. If the hypothese turns out to be true, you can get the performance improvement results in a quantified manner using the profilers itself.
You can find out the functions which are expensive (typically having a high value of the product of "Number of calls" and "Execution time per call"), and explore design decisions like adding a cache (if the input domain of the function is limited). You can go down a level deeper and use a line profiler to figure out what lines inside the function are the most time consuming and try to see if there are alternative ways.
However, synthetic benchmarks aren't enough. Sometimes things can vary a lot between production and development environment. Profiling in production is helpful. However, though, in most of the cases, we would not want to bear the overhead of operating a deterministic profiler in the production environment.
Following are some of the easy to fix ideas that developers often come across while profiling for CPU performance.
This the most obvious low-hanging fruit. Synchronous waiting in any form (time.sleep, thread.wait, lock.acquire, etc.) will block your program to execute anything on CPU. This blocked CPU time can better be utilized by other areas of your program, which don't depend on the wait time.
When your program is waiting for I/O, it is not utilizing the CPU. Some examples include disk reads, database queries, network communication (RPCs, REST API calls, etc.), etc.
Since logging to disk (or screen) is also an I/O bound task, you should be cautious about the things you're logging and the logging levels in production.
The work around with I/O bound taks and time delays is to execute them asynchronously (see asyncio for python3).
Every function call has an overhead. While profiling, you can try to classify expensive function calls as
- Genuine calls
- Un-necessary calls (alogrithmic issues)
This can be done using caller and callee info; time spent info and profile visualizations.
In case of genuine calls, you might want to explore implementing your function at a lower level (using cTypes, or Cython, or C) or use the libraries that already do so. For instance matrix calculations are much-much faster with Numpy than with native Python because numpy is built in CPython and uses SIMD architecture. Similarly, there are libraries like pandas, scipy, pycrypto, lxml, gstreamer, etc. which you can use for doing specific things instead of doing them on your own. There's also another generic suggestion that is hidden here "Don’t reinvent the wheel, look for existing stuff first".
There are some object conversion tasks that are CPU intensive and might not be necessary for your use-case. Examples include,
- Serialization
- Encoding & decoding, Encryption & decryption
- Compression and decompression
- Un-necessary conversion from one class to another
There are multiple ways to do the same things in Python, and their performance impact can differ drastically. Here are a few instances where you should be careful about performance impacts,
- String concatenation
- Naive string parsing, and using regexes
- Loading huge data in RAM
Parallelizing expensive tasks is also a good idea for optimizing performance. As you might know already, multithreading in python will provide concurrency and not actual parallelization. For that, you can use the multiprocessing module. Ideally, you can spawn upto as many as 1.5 times the number of cores processes (you might need to tweak this ratio a bit to find the "sweet-spot" for your application). Creating more processes also helps if your program uses cooperative multithreading (e.g., Greenlets in gevent). You can also explore the feasiblity of using tools like Dask, and PySpark (see our post on distributed machine learning with pyspark).
Heisenberg's uncertainty principle in quantum mechanics states that the more precisely you know the position of something, the less precisely you can tell its speed and momentum. A similar parallel can be drawn when it comes to profiling, i.e., by measuring one thing closely, you have actually weakened your understanding of the other. You have your code operating one way, and you try to measure it really deeply, put tons of hooks into it to do this monitoring using profilers, and it might actually change the performance of your code. So the final numbers may look somewhat different. Not even the code injected with profiler hooks will run x% slower, this x% might not be evenly distributed as well. So if you want high-precision or you're looking at the percentage-based metrics, this is something you should keep in mind.
- The profiling results might depend on external circumstances (Load avg on the machine, network speed, etc.). So profiling with an intent to compare two programs, you should try to keep the external conditions as much similar as possible.
Some tools might support limited versions of Python or only specific operating systems.
For instance, a statistical profiler is less likely to distort timings of a program that has a lot of function calls (like a Django template renderer). It also means that the profiler misses recording information about some of the function frames. However, the missed function frames are the fast running ones. This might be an acceptable tradeoff in the production environment since the overhead is very less and profilers are expected to find the slowest code. But in the development environment, we will want to have the definite answers to everything, so we should use deterministic profiler there. Similar tradeoffs exist when it comes to visualization, report formats, etc.
The are also some side-effects of optimization.
- Writing optimized code is slow. It requires a lot of human-effort (and that too experts) and time. Examples include writing computing libraries like numpy and tensorflow.
- Optimization can compromise with readablity. An optimized code will raise the bar for the level of expertise that a programmer should have to be able to understand it. Depending on the use-case you might have to end up writing code in lower level languages C, Cython, CUDA, etc.
- Optimization can also compromise with extensiblity / maintainablity. Some optimizations like Caching, lazy execution, parallelized executions, and hacky-patches are based on assumptions that things will be behaving in a certain way. And if the requirements on which these assumptions are based change in the near future, the code needs to be changed to account for that. A couple of examples can be,
- Caching becoming ineffective due to change in assumptions
- The side-effects of problems like race condition may suddendly become evident in case of high concurrent scale.
- The space-time tradeoff that you took in your design to improve the speed has somehow become a bottleneck now.
Writing code that does the job is one thing; writing it in an optimized way is a different game altogether. It not only requires the familiarity with differenet programming language constructs, but it also demands in-depth knowledge of how things work under the hood. Of course, there are tools and tips and tricks out there to help you optimize the code. But trust me, having a better picture of what's happening at a lower level, will help you go a long way in writing optimized code in the first place.
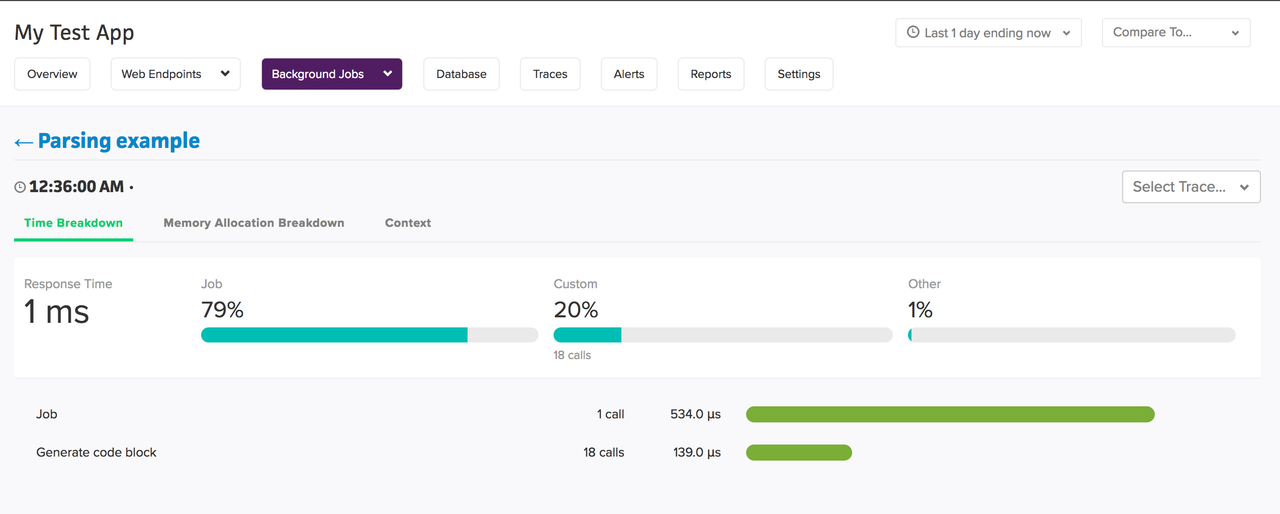
A good APM actively monitors different parts of your applications and can help you locate the hotspots for performance improvements very quickly. An APM may not be a silver bullet for optimizing your application, but it's a collection of lead bullets that can help in figuring out bottlenecks. For instance, Scout provides,
We mostly covered tools specific to profiling execution speeds native to python, but your application can be much more than a program that runs on CPU. For instance, if you have a web application that serves decent traffic, it is highly likely that your stack will have components databases, external API integrations, consumers and producers, and so on. And sometimes, the hotspots for optimizations can be outside the python runtime. Though there are tools available to profile each ot these components individually, a good APM tool will provide you the big-picture-overview of the system and the capability to dig deeper into the components. It'll record information at different layers of the stack. This is particularly useful for large applications.
Scout works on the concept of transactions (and traces). So it is determisintic in the sense every single transaction under monitoring is measured. To collect statistics related to different components scout utilizes techniques like signals and events, patching, etc.
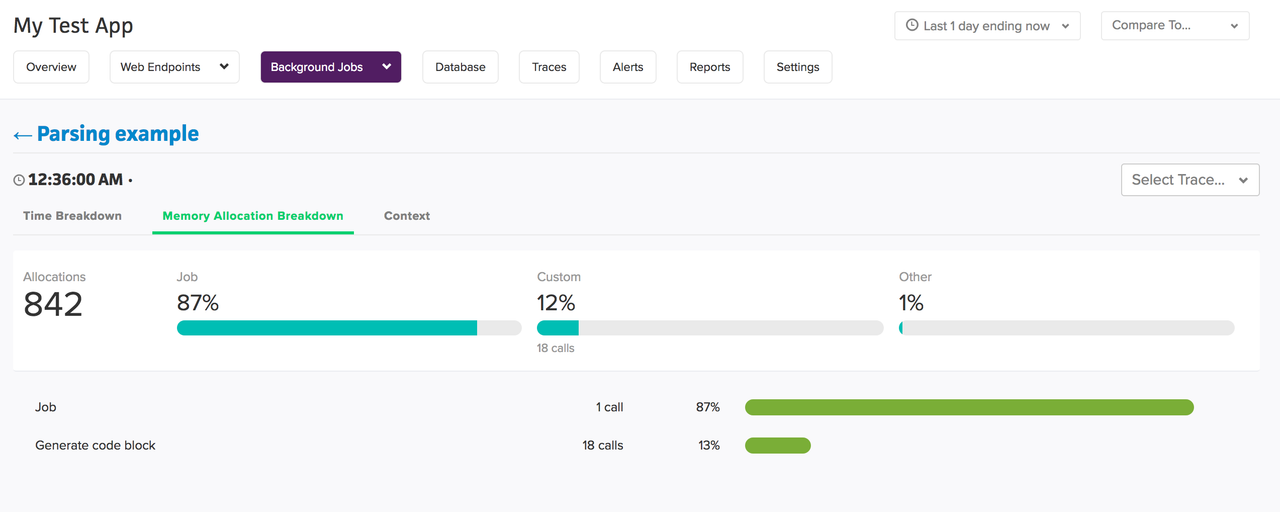
We didn't cover python-specific memory profilers in this post (since the post is already too long!), but scout has features like,
- Object allocation tracking (garbage collection)
- Ability to track process memory increases in RAM and tie them to specific transactions.
You can attach meaningful context to transactions in Scout to help correlate and identify performance problems to a specific context (users, actions, etc.). Scout ties each transaction to the Git version and will point you directly to the line of code responsible. Scout can aggregate results for various machines in your cluster.
If you have a web application running on a python web framework like Django or Flask, you can integrate flask right out-of-the box with a few lines of configuration.
Scout Usage
$ pip install scout-apmIf you have a flask, django, bottle, falcon, or pyramid application you can configure scout in a few lines and get moving.
In Django
# settings.py
INSTALLED_APPS = [
"scout_apm.django", # should be listed first
# ... other apps ...
]
# Scout settings
SCOUT_MONITOR = True
SCOUT_KEY = "[AVAILABLE IN THE SCOUT UI]"
SCOUT_NAME = "A FRIENDLY NAME FOR YOUR APP."In flask
from scout_apm.flask import ScoutApm
# Setup a flask 'app' as normal
# Attach ScoutApm to the Flask App
ScoutApm(app)
# Scout settings
app.config["SCOUT_MONITOR"] = True
app.config["SCOUT_KEY"] = "[AVAILABLE IN THE SCOUT UI]"
app.config["SCOUT_NAME"] = "A FRIENDLY NAME FOR YOUR APP"Scout will automatically instrument code related to following libraries,
- PyMongo
- Requests
- UrlLib3
- Redis
- ElasticSearch
- Jinja2
To monitor something else, you can use custom instrumentation. You can use the @scout_apm.api.BackgroundTransaction decorator or with scout_apm.api.BackgroundTransaction to specify the portion of the code you want to monitor, and if within that code if you want to measure a specific piece of work, you can use @scout_apm.api.instrument() decorator or with with scout_apm.api.instrument context manager for that,
import scout_apm.api
config = {'name': 'My Test App',
'key': 'MY_SCOUT_KEY',
'monitor': True}
scout_apm.api.install(config=config)
@scout_apm.api.BackgroundTransaction("Some name")
def some_func():
# some work
sub_func()
# some more work
@scout_apm.api.instrument("Sub function")
def sub_func():
# some workIn this post, we went-through different tools and techniques to identify performance bottlenecks in a python application, and some ideas to optimize performance of the application. We discussed about different profiling and visualizing tools available for profiling CPU performance in python, and their typical usage. We briefly discussed about the approach that we should follow when trying to optimize and some caveats associated with profiling and optimization. At last, we had a look at some of the features of Scout that can be helpful during profiling large applications.