Created
January 25, 2023 14:00
-
-
Save seandavi/b729199db0fdebe57029bdc85259f892 to your computer and use it in GitHub Desktop.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| --- | |
| title: "data-engineering-R" | |
| --- | |
| ## Background^[From https://www.stitchdata.com/columnardatabase/] | |
| Suppose you're a retailer maintaining a web-based storefront. An ecommerce site generates a lot of data. Consider product purchase transactions: | |
| 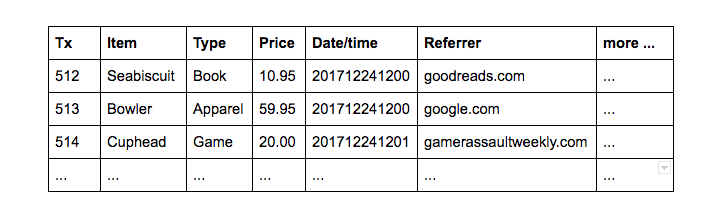 | |
| Businesses handle transactions using online transaction-processing (OLTP) software. All the fields in each row are important, so for OLTP it makes sense to store items on disk by row, with each field adjacent to the next in the same block on the hard drive: | |
| ``` | |
| 512,Seabiscuit,Book,10.95,201712241200,goodreads.com | |
| 513,Bowler,Apparel,59.95,201712241200,google.com | |
| 514.Cuphead,Game,20.00,201712241201,gamerassaultweekly.com | |
| ``` | |
| Transaction data is also characterized by frequent writes of individual rows. | |
| Some — but not all — of the information from transactions is useful to inform business decisions – what's called online analytical processing (OLAP). For instance, a retailer might want see how price affects sales, or to zero in on the referrers that send it the most traffic so it can determine where to advertise. For queries like these, we don't care about row-by-row values, but rather the information in certain columns for all rows. | |
| For OLAP purposes, it's better to store information in a columnar database, where blocks on the disk might look like: | |
| ``` | |
| 512,513,514 | |
| Seabiscuit,Bowler,Cuphead | |
| Book,Apparel,Game | |
| 10.95,59.95,20.00 | |
| 201712241200,201712241200,201712241201 | |
| goodreads.com,google.com,gamerassaultweekly.com | |
| ``` | |
| With this organization, applications can read the kinds of information you might want to analyze — pricing information, or referrerers — together in a single block. You get performance wins both by retrieving information that's grouped together, and by not retrieving information you don't need, such as individual names. | |
| ## Getting started | |
| ```{r eval=FALSE} | |
| BiocManager::install('duckdb') | |
| ``` | |
| ```{r hide=TRUE} | |
| unlink('my-db.duckdb') | |
| ``` | |
| The documentation for the client API for R is [available online](https://duckdb.org/docs/api/r). Note that the `install.packages` above brings along with it the DuckDB engine as well as the R client. Therefore, DuckDB is known as an __embedded__ database. The most popular embedded database is SQLite and DuckDB shares some similarities. | |
| * Pros of embedded databases | |
| * Small | |
| * Usually well-supported across systems | |
| * Easy to install and maintain | |
| * Data storage is often in one file or self-contained directory, making data sharing relatively easy | |
| * Cons of embedded databases | |
| * Limited in scalability | |
| * Usually not good for "high concurrency" | |
| * Not a client-server model, so data access must be from the host system (no remote connections) | |
| ```{r} | |
| library(duckdb) | |
| ``` | |
| The DuckDB library, like other SQL libraries in R, does not supply direct access functions to the database. Instead, two other libraries supply two different access APIs to the underlying database. | |
| The `DBI` package is the older package and supplies a set of generic functions for interacting with databases. The `dbplyr` package is newer and adapts the tidyverse and `dplyr` API to work with SQL databases. Both methods ultimately translate queries into SQL that is sent to the database. | |
| ## DBI^[Adapted from the DuckDB documentation for the R client] | |
| To use DuckDB, you must first create a connection object that represents the database. The connection object takes as parameter the database file to read and write from. If the database file does not exist, it will be created (the file extension may be `.db`, `.duckdb`, or anything else). The special value `:memory:` (the default) can be used to create an **in-memory database**. Note that for an in-memory database no data is persisted to disk (i.e. all data is lost when you exit the R process). If you would like to connect to an existing database in read-only mode, set the `read_only` flag to `TRUE`. _Read-only mode is required if multiple R processes want to access the same database file at the same time_. | |
| ```{r} | |
| library("DBI") | |
| # to start an in-memory database | |
| con <- dbConnect(duckdb::duckdb(), dbdir = ":memory:") | |
| # to use a database file (not shared between processes) | |
| con <- dbConnect(duckdb::duckdb(), dbdir = "my-db.duckdb", read_only = FALSE) | |
| ``` | |
| Connections are closed implicitly when they go out of scope or if they are explicitly closed using `dbDisconnect()`. To shut down the database instance associated with the connection, use `dbDisconnect(con, shutdown=TRUE)` | |
| ### [Querying](https://duckdb.org/docs/api/r#querying) | |
| DuckDB supports the standard DBI methods to send queries and retrieve result sets. `dbExecute()` is meant for queries where no results are expected like `CREATE TABLE` or `UPDATE` etc. and `dbGetQuery()` is meant to be used for queries that produce results (e.g. `SELECT`). Below an example. | |
| ```{r} | |
| # create a table | |
| dbExecute(con, "CREATE TABLE items(item VARCHAR, value DECIMAL(10,2), count INTEGER)") | |
| ``` | |
| The table is called `items` and has two columns: | |
| 1. `item`, of type VARCHAR (variable length character, what we would call a "string") | |
| 2. `value`, of type DECIMAL (with set precision) | |
| 3. `count`, of type INTEGER | |
| To delete the same table: | |
| ```{r} | |
| dbExecute(con, "DROP TABLE items") | |
| ``` | |
| We can also replace a table with `CREATE OR REPLACE TABLE`. | |
| ```{r} | |
| # create OR REPLACE a table | |
| dbExecute(con, "CREATE OR REPLACE TABLE items(item VARCHAR, value DECIMAL(10,2), count INTEGER)") | |
| ``` | |
| The `CREATE TABLE` statement _just_ creates the table, but nothing is in it. | |
| ```{r} | |
| # insert two items into the table | |
| dbExecute(con, "INSERT INTO items VALUES \ | |
| ('jeans', 20.0, 1), \ | |
| ('hammer', 42.2, 2)") | |
| ``` | |
| ```{r} | |
| # retrieve the items again | |
| res <- dbGetQuery(con, "SELECT item, value, count FROM items") | |
| print(res) | |
| ``` | |
| It one wants to select _all_ columns, there is a shortcut: | |
| ```{r} | |
| res <- dbGetQuery(con, "SELECT * FROM items") | |
| print(res) | |
| ``` | |
| DuckDB also supports _prepared statements_ in the R API with the `dbExecute` and `dbGetQuery` methods. Prepared statements allow the database to develop the "plan" for execution only once and then to reuse that plan. In other words, for queries that are going to be reused, prepared statements can be more efficient. Here are a few examples, though in these examples, there probably isn't any measurable difference in speed. | |
| ```{r} | |
| # prepared statement parameters are given as a list | |
| dbExecute(con, "INSERT INTO items VALUES (?, ?, ?)", list('laptop', 2000, 1)) | |
| # if you want to reuse a prepared statement multiple times, use dbSendStatement() and dbBind() | |
| stmt <- dbSendStatement(con, "INSERT INTO items VALUES (?, ?, ?)") | |
| dbBind(stmt, list('iphone', 300, 2)) | |
| dbBind(stmt, list('android', 3.5, 1)) | |
| dbClearResult(stmt) | |
| # query the database using a prepared statement | |
| res <- dbGetQuery(con, "SELECT item FROM items WHERE value > ?", list(400)) | |
| print(res) | |
| ``` | |
| > #### Note | |
| > | |
| > Do **not** use prepared statements to insert large amounts of data into DuckDB. See below for better options. | |
| ## [Efficient Transfer](https://duckdb.org/docs/api/r#efficient-transfer) | |
| To write a R data frame into DuckDB, use the standard DBI function `dbWriteTable()`. This creates a table in DuckDB and populates it with the data frame contents. For example: | |
| ```{r} | |
| dbWriteTable(con, "iris_table", iris) | |
| res <- dbGetQuery(con, "SELECT * FROM iris_table LIMIT 3") | |
| print(res) | |
| ``` | |
| A slightly more interesting example: | |
| ```{r} | |
| res <- dbGetQuery(con, "SELECT \ | |
| species, count(*) \ | |
| FROM \ | |
| iris_table \ | |
| GROUP BY species") | |
| print(res) | |
| ``` | |
| It is also possible to “register” a R data frame as a virtual table, comparable to a SQL `VIEW`. This _does not actually transfer data_ into DuckDB yet. Below is an example: | |
| ``` | |
| duckdb::duckdb_register(con, "iris_view", iris) | |
| res <- dbGetQuery(con, "SELECT * FROM iris_view LIMIT 1") | |
| print(res) | |
| ``` | |
| > #### Note | |
| > | |
| > DuckDB keeps a reference to the R data frame after registration. This prevents the data frame from being garbage-collected. The reference is cleared when the connection is closed, but can also be cleared manually using the `duckdb::duckdb_unregister()` method. | |
| Also refer to [the data import documentation](https://duckdb.org/docs/api/r../data/overview) for more options of efficiently importing data. | |
| ## [dbplyr](https://duckdb.org/docs/api/r#dbplyr) | |
| DuckDB also plays well with the [dbplyr](https://CRAN.R-project.org/package=dbplyr) / [dplyr](https://dplyr.tidyverse.org) packages for programmatic query construction from R. Here is an example: | |
| ```{r} | |
| library("DBI") | |
| library("dplyr") | |
| con <- dbConnect(duckdb::duckdb()) | |
| duckdb::duckdb_register(con, "flights", nycflights13::flights) | |
| tbl(con, "flights") |> | |
| group_by(dest) |> | |
| summarise(delay = mean(dep_time, na.rm = TRUE)) |> | |
| collect() | |
| ``` | |
| When using dbplyr, CSV and Parquet files can be read using the `dplyr::tbl` function. | |
| ```{r} | |
| write.csv(mtcars, "mtcars.csv") | |
| tbl(con, "mtcars.csv") |> | |
| group_by(cyl) |> | |
| summarise(across(disp:wt, .fns = mean)) |> | |
| collect() | |
| ``` | |
| ## Working with Parquet | |
| ```{r message=FALSE} | |
| taxi_file = 'taxi_2019_04.parquet' | |
| download.file('https://github.com/cwida/duckdb-data/releases/download/v1.0/taxi_2019_04.parquet', taxi_file) | |
| file.size(taxi_file) | |
| ``` | |
| What's in here? | |
| ```{r} | |
| dbGetQuery(con, "select count(*) from 'taxi_2019_04.parquet'") | |
| ``` | |
| ```{r} | |
| res = dbGetQuery(con, "select * from 'taxi_2019_04.parquet' limit 5") | |
| print(res) | |
| ``` | |
| ```{r} | |
| res = dbGetQuery(con, "SELECT dropoff_location_id, count(*)\ | |
| FROM 'taxi_2019_04.parquet' \ | |
| GROUP BY dropoff_location_id | |
| ORDER BY count(*) desc | |
| LIMIT 10") | |
| print(res) | |
| ``` | |
| ## Remote files (s3, GCS, http(s)) | |
| ```{r} | |
| dbExecute(con, "install 'httpfs'") | |
| dbExecute(con, "load 'httpfs'") | |
| ``` | |
| ```{r} | |
| dbGetQuery(con, "select * from 'https://github.com/cwida/duckdb-data/releases/download/v1.0/taxi_2019_04.parquet' limit 10") | |
| ``` | |
| ```{r} | |
| res = dbGetQuery(con, "SELECT dropoff_location_id, count(*)\ | |
| FROM \ | |
| 'https://github.com/cwida/duckdb-data/releases/download/v1.0/taxi_2019_04.parquet' \ | |
| GROUP BY dropoff_location_id | |
| ORDER BY count(*) desc | |
| LIMIT 10") | |
| print(res) | |
| ``` | |
| ```{r} | |
| res = dbGetQuery(con, "SELECT dropoff_location_id, count(*)\ | |
| FROM \ | |
| 'https://github.com/cwida/duckdb-data/releases/download/v1.0/taxi_2019_04.parquet' \ | |
| WHERE pickup_at BETWEEN '2019-04-15' AND '2019-04-20' \ | |
| GROUP BY dropoff_location_id | |
| ORDER BY count(*) desc | |
| LIMIT 10") | |
| print(res) | |
| ``` | |
| ```{r} | |
| res = dbGetQuery(con, "SELECT dropoff_location_id, count(*)\ | |
| FROM \ | |
| parquet_scan([\ | |
| 'https://github.com/cwida/duckdb-data/releases/download/v1.0/taxi_2019_04.parquet',\ | |
| 'https://github.com/cwida/duckdb-data/releases/download/v1.0/taxi_2019_05.parquet',\ | |
| 'https://github.com/cwida/duckdb-data/releases/download/v1.0/taxi_2019_06.parquet' | |
| ]) \ | |
| WHERE pickup_at BETWEEN '2019-05-15' AND '2019-05-20' \ | |
| GROUP BY dropoff_location_id | |
| ORDER BY count(*) desc | |
| LIMIT 10") | |
| print(res) | |
| ``` |
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment