output:
rmarkdown::html_document:
highlight: pygments
toc: true
toc_depth: 3
fig_width: 5
bibliography: "r system.file(package='dummychapter1', 'vignettes', 'bibliography.bib')"
vignette: >
%\VignetteIndexEntry{dummychapter1}
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| # <seandavi@gmail.com>, 2023-06-02 | |
| # | |
| # Terraform for setting up GPT-4, GPT-3.5-turbo, and text-embedding-ada-002 | |
| # endpoints. Note that not all models are available in all regions, so | |
| # check before changing the region here, currently set to "southcentralus" | |
| # | |
| # Assumes az-cli authenticated (requires Azure subscription) and terraform | |
| # available and installed | |
| # | |
| terraform { |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| --- | |
| title: "data-engineering-R" | |
| --- | |
| ## Background^[From https://www.stitchdata.com/columnardatabase/] | |
| Suppose you're a retailer maintaining a web-based storefront. An ecommerce site generates a lot of data. Consider product purchase transactions: | |
| 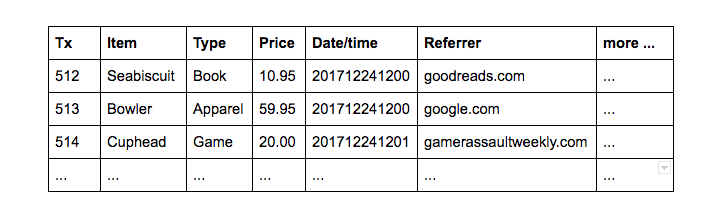 |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| {"title":"Mesothelioma_52413","status":"Public on Dec 23 2022","submission_date":"2022-08-05","last_update_date":"2022-12-23","type":"genomic","anchor":null,"contact":{"city":"Nagoya","name":{"first":"Shinya","middle":"","last":"Toyokuni"},"email":"akatsuka@med.nagoya-u.ac.jp","state":"Aichi","address":"65 Tsuruma-Cho, Showa-Ku","department":"Pathology","country":"Japan","web_link":null,"institute":"Nagoya University","zip_postal_code":null,"phone":null},"description":null,"accession":"GSM6433302","biosample":null,"tag_count":null,"tag_length":null,"platform_id":"GPL10451","hyb_protocol":"The labeled DNA was hybridized with Agilent SurePrint G3 Mouse CGH 4x180k microarray at 67°C for 24 hours according to the manufacturer's protocol (Version 8.0).","channel_count":2,"scan_protocol":"The slides were scanned in an Agilent DNA microarray scanner with SureScan High-Resolution Technology (G2565CA).","data_row_count":174012,"library_source":null,"overall_design":null,"sra_experiment":null,"data_processing":"The sc |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| ## ----------------------------------------------------------------------------- | |
| ## GEOquery | |
| ## ----------------------------------------------------------------------------- | |
| library(GEOquery) | |
| gse = getGEO("GSE103512")[[1]] | |
| ## ----------------------------------------------------------------------------- | |
| library(SummarizedExperiment) | |
| se = as(gse, "SummarizedExperiment") |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| ## ----message=FALSE,warning=FALSE---------------------------------------------- | |
| pkgs = c( | |
| "ggplot2", | |
| "GEOquery", | |
| "SummarizedExperiment" | |
| ) | |
| ins = installed.packages(repos = BiocManager::repositories()) | |
| for(pkg in pkgs) { | |
| if(!(pkg %in% rownames(ins))) | |
| BiocManager::install(pkg) |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| #!/bin/bash | |
| # requires about 200G of disk space | |
| # downloads stuff | |
| # create disposable bucket | |
| # upload | |
| # bq load | |
| # remove bucket | |
| mkdir -p ss | |
| cd ss | |
| wget https://s3-us-west-2.amazonaws.com/ai2-s2-research-public/open-corpus/2022-01-01/manifest.txt |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| #!/bin/bash | |
| CONTAINER=ghcr.io/seandavi/buildabiocworkshop | |
| ZONE=us-central1-a | |
| PASSWORD=rstudio | |
| INSTANCE=rs-2 | |
| gcloud compute instances create-with-container $INSTANCE \ | |
| --container-image $CONTAINER \ | |
| --container-env PASSWORD=$PASSWORD \ | |
| --tags rstudio |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| #!/bin/bash | |
| gcloud compute instances create myinstance \ | |
| --metadata-from-file=startup-script=startup.sh \ | |
| --scopes=compute-rw |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| start_date='2000-01-01' | |
| end_date = '2021-12-31' | |
| datefilter = function(date) { | |
| startdate = format(date,'%Y-%m-%d') | |
| return(sprintf("dt:release:from=%suntil=%s",startdate,startdate)) | |
| } | |
| download_biosample = function(date) { | |
| require(httr) |