Created
August 21, 2023 13:55
-
-
Save simonrolph/4096eeb9dc230539193e0161d87710d6 to your computer and use it in GitHub Desktop.
R targets workflow with static branches per species and combined trend analysis
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| # This is an example r {targets} workflow it does the following: | |
| # | |
| # (0. (Generate some example data) | |
| # 1. Statically branch to create data subsets for each species | |
| # 2. Fit a linear model to each of the species subset | |
| # 3. Produce and save a plot of each species trend (with a line from the linear model) using tar_map() | |
| # 4. Produce and save a plot with all of the species combined and their linear models using tar_combine() | |
| #this is combined into a single file for sharability but normally you'd have the R functions in another .R file | |
| #R 4.2.2, targets_1.2.2 and tarchetypes_0.7.7 | |
| #run interactively to generate example dataset - then don't run again | |
| if(F){ #don't run when doing tar_make() | |
| species = c("vulpes_vulpes","bufo_bufo", "plantago_lanceolata","plantago_major") | |
| abundance <- expand.grid(abundance = 100:80 , species = species) | |
| abundance$time <- 1990:2010 | |
| abundance$abundance <- abundance$abundance - sample(1:20, size = nrow(abundance),replace=T) | |
| write.csv(abundance,"abundance.csv",row.names = F) | |
| glimpse(abundance) | |
| } | |
| #---------- | |
| #load packages | |
| library(targets) | |
| library(tarchetypes) #required for tar_map() | |
| #define your focal species, might not be all the species in the dataset (for the static branching) | |
| interested_species <- data.frame(foc_species = c("plantago_lanceolata","plantago_major","bufo_bufo")) | |
| ### targets functions | |
| # a function to subset for a certain condition eg. a species | |
| subset_data <- function(dataset,focal_species){ | |
| dataset[dataset$species == focal_species,] | |
| } | |
| # a function for fitting a model | |
| fit_model <- function(dataset){ | |
| lm(abundance~time,dataset) | |
| } | |
| # a function for plotting points, and adding lm line from model | |
| # will be format="file" | |
| plot_with_line <- function(dataset,model,species_name){ | |
| file_name <- paste0("plot_",species_name,".png") | |
| png(file=file_name,width=600, height=600) | |
| plot(dataset$time,dataset$abundance,main = species_name) | |
| abline(model) | |
| dev.off() | |
| #return just the file name | |
| file_name | |
| } | |
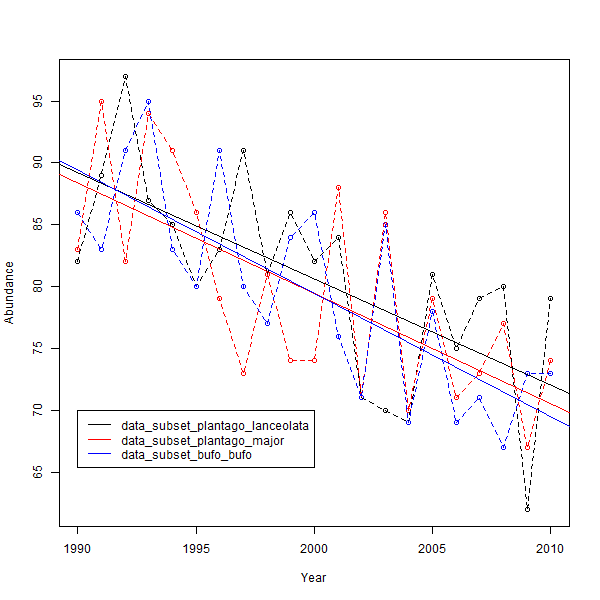
| plot_combined <- function(models,data_subsets){ | |
| file_name <- "plot_combined.png" | |
| png(file=file_name,width=600, height=600) | |
| #add the datasets | |
| plot(data_subsets[[1]]$time,data_subsets[[1]]$abundance,xlab = "Year",ylab="Abundance") | |
| points(data_subsets[[2]]$time,data_subsets[[2]]$abundance,col = "red") | |
| points(data_subsets[[3]]$time,data_subsets[[3]]$abundance,col = "blue") | |
| #join the dots | |
| lines(data_subsets[[1]]$time,data_subsets[[1]]$abundance,lty=2) | |
| lines(data_subsets[[2]]$time,data_subsets[[2]]$abundance,col = "red",lty=2) | |
| lines(data_subsets[[3]]$time,data_subsets[[3]]$abundance,col = "blue",lty=2) | |
| #add the lines from the models | |
| abline(models[[1]]) | |
| abline(models[[2]],col = "red") | |
| abline(models[[3]],col = "blue") | |
| #add a legend | |
| legend(1990, 70, legend=names(data_subsets)[1:3], | |
| col=c("black","red", "blue"),lty=1) | |
| dev.off() | |
| #return just the file name | |
| file_name | |
| } | |
| ### targets workflow | |
| #first we need to define sub workflows split into data files for each species | |
| mapped <- tar_map( | |
| #map over the interested species dataframe | |
| values = interested_species, | |
| #each thing we do for all species | |
| tar_target(data_subset, subset_data(data,foc_species)), #subset to only that species | |
| tar_target(subset_model, fit_model(data_subset)), #fit a model | |
| tar_target(out_plot,plot_with_line(data_subset,subset_model,foc_species)) # make a plot | |
| ) | |
| #built the workflow list | |
| list( | |
| tar_target(data_file, "abundance.csv",format="file"), | |
| tar_target(data ,read.csv(data_file)), | |
| # sub workflow defined above for each species | |
| mapped, | |
| #first combine them to lists of each data type simply using the list() function | |
| tar_combine(models_list,mapped$subset_model,command = list(!!!.x)), | |
| tar_combine(data_list,mapped$data_subset,command = list(!!!.x)), | |
| #then make a plot using the normal tar_target functions using the combined targets | |
| tar_target(combined_plot,plot_combined(models_list,data_list),format = "file") | |
| ) | |
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment

Final plot should look something like this