guppy_basecaller --disable_pings --compress_fastq -c dna_r9.4.1_450bps_fast.cfg -i flongle_fast5_pass/ -s flongle_test2 -x 'auto' --recursive guppy_basecaller --disable_pings --compress_fastq -c dna_r9.4.1_450bps_modbases_dam-dcm-cpg_hac.cfg --fast5_out -i flongle_fast5_pass/ -s flongle_hac_fastq -x 'auto' --recursive $ guppy_basecaller --compress_fastq -c dna_r9.4.1_450bps_modbases_dam-dcm-cpg_hac.cfg -i flongle_fast5_pass/ -s flongle_hac_fastq -x 'auto' --recursive
ONT Guppy basecalling software version 3.4.1+213a60d0
config file: /opt/ont/guppy/data/dna_r9.4.1_450bps_modbases_dam-dcm-cpg_hac.cfg
model file: /opt/ont/guppy/data/template_r9.4.1_450bps_modbases_dam-dcm-cpg_hac.jsn
input path: flongle_fast5_pass/
save path: flongle_hac_fastq
chunk size: 1000
chunks per runner: 512
records per file: 4000
fastq compression: ON
num basecallers: 1
gpu device: auto
kernel path:
runners per device: 4
Found 105 fast5 files to process.
Init time: 2790 ms
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
Caller time: 2493578 ms, Samples called: 3970728746, samples/s: 1.59238e+06
Finishing up any open output files.
Basecalling completed successfully.
So from the above we see in high accuracy mode it take the Xavier ~41 minutes to complete the base calling using the default configuration files. For reference the fast calling mode was ~8 minutes.
- When performing GPU basecalling there is always one CPU support thread per GPU caller, so the number of callers (
--num_callers) dictates the maximum number of CPU threads used. - Max chunks per runner (
--chunks_per_runner): The maximum number of chunks which can be submitted to a single neural network runner before it starts computation. Increasing this figure will increase GPU basecalling performance when it is enabled. - Number of GPU runners per device (
--gpu_runners_per_device): The number of neural network runners to create per CUDA device. Increasing this number may improve performance on GPUs with a large number of compute cores, but will increase GPU memory use. This option only affects GPU calling.
There is a rough equation to estimate amount of ram:
runners * chunks_per_runner * chunk_size < 100000 * [max GPU memory in GB]
For example, a GPU with 8 GB of memory would require:
runners * chunks_per_runner * chunk_size < 800000
--num_callers 1
--gpu_runners_per_device 2
--chunks_per_runner 48
chunk_size = 1000
gpu_runners_per_device = 4
chunks_per_runner = 512
chunks_per_caller = 10000
guppy_basecaller --disable_pings --compress_fastq -c dna_r9.4.1_450bps_fast.cfg -i flongle_fast5_pass/ \
-s flongle_test2 -x 'auto' --recursive --num_callers 4 --gpu_runners_per_device 8 --chunks_per_runner 256$ guppy_basecaller --disable_pings --compress_fastq -c dna_r9.4.1_450bps_fast.cfg \
-i flongle_fast5_pass/ -s flongle_test2 -x 'auto' --recursive --num_callers 4 \
--gpu_runners_per_device 8 --chunks_per_runner 256
ONT Guppy basecalling software version 3.4.1+213a60d0
config file: /opt/ont/guppy/data/dna_r9.4.1_450bps_fast.cfg
model file: /opt/ont/guppy/data/template_r9.4.1_450bps_fast.jsn
input path: flongle_fast5_pass/
save path: flongle_test2
chunk size: 1000
chunks per runner: 256
records per file: 4000
fastq compression: ON
num basecallers: 4
gpu device: auto
kernel path:
runners per device: 8
Found 105 fast5 files to process.
Init time: 880 ms
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
Caller time: 428745 ms, Samples called: 3970269916, samples/s: 9.26021e+06
Finishing up any open output files.
Basecalling completed successfully.
I was able to shave a minute off the fast model on the Xavier (above) getting it down to ~7 minutes.
Update: (13th Dec 2019)
Just modifying the number of chunks per runner has allowed me to get the time down to under 6.5 mins (see table below).
| chunks_per_runner | time |
|---|---|
| (160) default | ~8 mins |
| 256 | 7 mins 6 secs |
| 512 | 6 mins 28 secs |
| 1024 | 6 min 23 secs |
It looks like we might have reached an optimal point here. Next I'll test some of the other parameters and see if we can speed this up further.
guppy_basecaller --disable_pings --compress_fastq -c dna_r9.4.1_450bps_modbases_dam-dcm-cpg_hac.cfg \
--num_callers 4 --gpu_runners_per_device 8 --fast5_out -i flongle_fast5_pass/ \
-s flongle_hac_basemod_fastq -x 'auto' --recursive$ guppy_basecaller --disable_pings --compress_fastq -c dna_r9.4.1_450bps_fast.cfg \
-i flongle_fast5_pass/ -s flongle_test2 -x 'auto' --recursive --num_callers 8 \
--gpu_runners_per_device 8 --chunks_per_runner 1024
ONT Guppy basecalling software version 3.4.1+213a60d0
config file: /opt/ont/guppy/data/dna_r9.4.1_450bps_fast.cfg
model file: /opt/ont/guppy/data/template_r9.4.1_450bps_fast.jsn
input path: flongle_fast5_pass/
save path: flongle_test2
chunk size: 1000
chunks per runner: 1024
records per file: 4000
fastq compression: ON
num basecallers: 8
gpu device: auto
kernel path:
runners per device: 8
Found 105 fast5 files to process.
Init time: 897 ms
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
Caller time: 383865 ms, Samples called: 3970269916, samples/s: 1.03429e+07
Finishing up any open output files.
Basecalling completed successfully.
$ guppy_basecaller --disable_pings --compress_fastq -c dna_r9.4.1_450bps_fast.cfg \
-i flongle_fast5_pass/ -s flongle_test2 -x 'auto' --recursive --num_callers 4 \
--gpu_runners_per_device 8 --chunks_per_runner 1024 --chunk_size 2000
ONT Guppy basecalling software version 3.4.1+213a60d0
config file: /opt/ont/guppy/data/dna_r9.4.1_450bps_fast.cfg
model file: /opt/ont/guppy/data/template_r9.4.1_450bps_fast.jsn
input path: flongle_fast5_pass/
save path: flongle_test2
chunk size: 2000
chunks per runner: 1024
records per file: 4000
fastq compression: ON
num basecallers: 4
gpu device: auto
kernel path:
runners per device: 8
Found 105 fast5 files to process.
Init time: 1180 ms
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
Caller time: 503532 ms, Samples called: 3970269916, samples/s: 7.88484e+06
Finishing up any open output files.
Basecalling completed successfully.
$ guppy_basecaller --disable_pings --compress_fastq -c dna_r9.4.1_450bps_fast.cfg \
-i flongle_fast5_pass/ -s flongle_test2 -x 'auto' --recursive --num_callers 8 \
--gpu_runners_per_device 16 --chunks_per_runner 1024 --chunk_size 1000
ONT Guppy basecalling software version 3.4.1+213a60d0
config file: /opt/ont/guppy/data/dna_r9.4.1_450bps_fast.cfg
model file: /opt/ont/guppy/data/template_r9.4.1_450bps_fast.jsn
input path: flongle_fast5_pass/
save path: flongle_test2
chunk size: 1000
chunks per runner: 1024
records per file: 4000
fastq compression: ON
num basecallers: 8
gpu device: auto
kernel path:
runners per device: 16
Found 105 fast5 files to process.
Init time: 1113 ms
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
Caller time: 383466 ms, Samples called: 3970269916, samples/s: 1.03536e+07
Finishing up any open output files.
Basecalling completed successfully.
The below parameters seem to provide the 'optimal' speed increase with a resultant run time of 6 mins and 23 secs.
$ guppy_basecaller --disable_pings --compress_fastq -c dna_r9.4.1_450bps_fast.cfg \
-i flongle_fast5_pass/ -s flongle_test2 -x 'auto' --recursive --num_callers 4 \
--gpu_runners_per_device 8 --chunks_per_runner 1024 --chunk_size 1000
ONT Guppy basecalling software version 3.4.1+213a60d0
config file: /opt/ont/guppy/data/dna_r9.4.1_450bps_fast.cfg
model file: /opt/ont/guppy/data/template_r9.4.1_450bps_fast.jsn
input path: flongle_fast5_pass/
save path: flongle_test2
chunk size: 1000
chunks per runner: 1024
records per file: 4000
fastq compression: ON
num basecallers: 4
gpu device: auto
kernel path:
runners per device: 8
Found 105 fast5 files to process.
Init time: 926 ms
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
Caller time: 382714 ms, Samples called: 3970269916, samples/s: 1.0374e+07
Finishing up any open output files.
Basecalling completed successfully.
We are currently using a 27000mAh AC Portable Charger from Ravpower.
Below: Ravpower Xtreme Series 27000mAh AC Portable Charger
 This battery bank/charger has a built in 220V AC outlet and 1 usb-c and 2 usb 3.1 outputs.
This battery bank/charger has a built in 220V AC outlet and 1 usb-c and 2 usb 3.1 outputs.
Below: powerbank charging from the wall.
 Ravpower claim this powerbank will charge a smartphone 11 times, a tablet 4 times or a laptop 3 times.
Ravpower claim this powerbank will charge a smartphone 11 times, a tablet 4 times or a laptop 3 times.
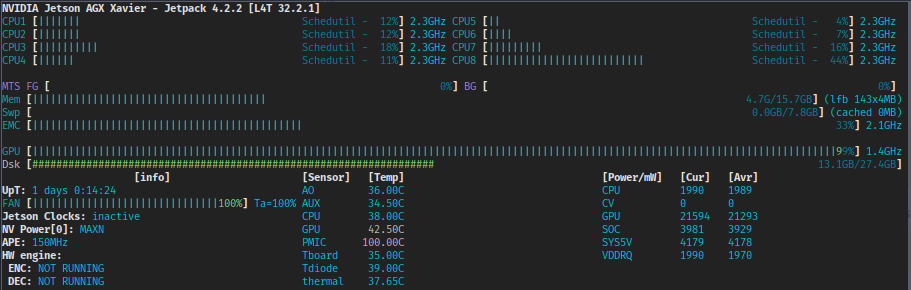
Below: running our first portable Xavier GPU basecalling of nanopore data!

Running the Xavier in different power states obviously influences the amount of run time on the battery.
| Power mode | Time |
|---|---|
| 10W | 33.4 mins |
| 15W | 14.3 mins |
| 30W 2 cores | 10.8 mins |
| 30W 4 cores | 10.8 mins |
| 30W MAX (8 cores) | 7.5 mins |
The above benchmarks were performed on data generated from a flongle run (~0.5 Mb of sequence or 5.5 Gb of actual data).
guppy_basecaller \
--disable_pings \
--compress_fastq \
-c dna_r9.4.1_450bps_modbases_dam-dcm-cpg_hac.cfg \
--ipc_threads 16 \
--num_callers 8 \
--gpu_runners_per_device 4 \
--chunks_per_runner 512 \
--device "cuda:0 cuda:1" \ # this parameter should now scale nicely across both cards, I haven't checked though
--recursive \
--fast5_out \
-i fast5_input \
-s fastq_output
guppy_basecaller \
--disable_pings \
--compress_fastq \
-c dna_r9.4.1_450bps_fast.cfg \
--ipc_threads 16 \
--num_callers 8 \
--gpu_runners_per_device 64 \
--chunks_per_runner 256 \
--device "cuda:0 cuda:1" \
--recursive \
-i fast5_input \
-s fastq_output
There has been some discussion about the recent release of Guppy (3.4.1 and 3.4.2) in terms of speed. I was interested in running some benchmarks across different versions. I had a hunch it may have been something to do with the newly introduced compression of the fast5 files...
The only things I am changing are the version of Guppy being used, and in the case of 3.4.3 I am trying with and without vbz compression of the fast5 files. Everything else is as below:
System:
- Debian Sid (unstable)
- 2x 12-Core Intel Xeon Gold 5118 (48 threads)
- 256Gb RAM
- Titan RTX
- Nvidia drivers: 418.56
- CUDA Version: 10.1
Guppy GPU basecalling parameters:
- --disable_pings
- --compress_fastq
- --dna_r9.4.1_450bps_fast.cfg
- --num_callers 8
- --gpu_runners_per_device 64
- --chunks_per_runner 256
- --device "cuda:0"
- --recursive
For each Guppy version I ran the basecaller three times in an attempt to ensure that results were consistent*.
Note: I chose the fast basecalling model as I wanted to do a quick set of benchmarks. If I feel up to it I may do the same thing for the high accuracy caller...
* Spoiler, I didn't originally do this and it proved misleading...
| guppy version | time (seconds) | samples/s |
|---|---|---|
| 3.1.5# | 93.278 | 4.25638e+07 |
| 3.2.4# | 94.141 | 4.21737e+07 |
| 3.3.0# | 94.953 | 4.1813e+07 |
| 3.3.3# | 95.802 | 4.14425e+07 |
| 3.4.1 (no vbz compressed fast5) | 79.913 | 4.96824e+07 |
| 3.4.1 (vbz compressed fast5) | 90.895 | 4.36797e+07 |
| 3.4.3 (no vbz compressed fast5) | 90.674 | 4.37862e+07 |
| 3.4.3 (vbz compressed fast5) | 82.877 | 4.79056e+07 |
# these versions of Guppy did not support vbz compression of fast5 files (pre 3.4.X from memory).
I initially thought that there was something off with the compression imlementation in 3,4,3 as my first run on uncompressed data was ~3x slower than the run on the compressed data. When I grabbed 3.4.1 to perform the same check I noticed that it was fairly consistent between compressed and not. So I went back and was more rigorous and performed 3 iterations of each run for each version, ditto for versions 3.4.X compressed and not. This proved that the initial run was an anomaly and should be disregarded.
What was quite interesting is that running on vbz compressed fast5 data appears to be in the range of 8-10 seconds faster than uncompressed. So there is a slight added speed benefit on top of the nice reduction in file size - which is a little nicer for the SSD/HDD.
So at this stage I can't confirm any detrimental speed issues when using Guppy version 3.4.X, but this needs to be caveated with all the usual disclaimers:
- all systems are different (I'm not on Ubuntu for instance).
- drivers are different (I need to update).
- GPUs are very different, i.e. many people (including me) are using 'non-supported' GPUs - in my case a Titan RTX which is no slouch.
- For what it's worth, I can add a comment here saying that I haven't had any speed issues with basecalling on our Nvidia Jetson Xaviers using Guppy 3.4.1.
- our 'little' Linux server isn't exactly a slouch either - so laptop/desktop builds could be very different.
- I ran the fast basecaller (I'm currently flat out and can't wait for the high accuracy caller) - I may take a subset of data and revist with hac at some stage.
You can view the 'raw' results/output for each run below:
~/Downloads/software/guppy/3.1.5/ont-guppy/bin/guppy_basecaller \
--disable_pings \
--compress_fastq \
-c dna_r9.4.1_450bps_fast.cfg \
--num_callers 8 \
--gpu_runners_per_device 64 \
--chunks_per_runner 256 \
--device "cuda:0" \
--recursive \
-i flongle_fast5_pass \
-s testrun_fast_3.1.5
ONT Guppy basecalling software version 3.1.5+781ed57
config file: /home/miles/Downloads/software/guppy/3.1.5/ont-guppy/data/dna_r9.4.1_450bps_fast.cfg
model file: /home/miles/Downloads/software/guppy/3.1.5/ont-guppy/data/template_r9.4.1_450bps_fast.jsn
input path: flongle_fast5_pass
save path: testrun_fast_3.1.5
chunk size: 1000
chunks per runner: 256
records per file: 4000
fastq compression: ON
num basecallers: 8
gpu device: cuda:0
kernel path:
runners per device: 64
Found 105 fast5 files to process.
Init time: 1000 ms
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
Caller time: 93278 ms, Samples called: 3970269916, samples/s: 4.25638e+07
Finishing up any open output files.
Basecalling completed successfully.
~/Downloads/software/guppy/3.2.4/ont-guppy/bin/guppy_basecaller \
--disable_pings \
--compress_fastq \
-c dna_r9.4.1_450bps_fast.cfg \
--num_callers 8 \
--gpu_runners_per_device 64 \
--chunks_per_runner 256 \
--device "cuda:0" \
--recursive \
-i flongle_fast5_pass \
-s testrun_fast_3.2.4
ONT Guppy basecalling software version 3.2.4+d9ed22f
config file: /home/miles/Downloads/software/guppy/3.2.4/ont-guppy/data/dna_r9.4.1_450bps_fast.cfg
model file: /home/miles/Downloads/software/guppy/3.2.4/ont-guppy/data/template_r9.4.1_450bps_fast.jsn
input path: flongle_fast5_pass
save path: testrun_fast_3.2.4
chunk size: 1000
chunks per runner: 256
records per file: 4000
fastq compression: ON
num basecallers: 8
gpu device: cuda:0
kernel path:
runners per device: 64
Found 105 fast5 files to process.
Init time: 836 ms
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
Caller time: 94141 ms, Samples called: 3970269916, samples/s: 4.21737e+07
Finishing up any open output files.
Basecalling completed successfully.
~/Downloads/software/guppy/3.3.0/ont-guppy/bin/guppy_basecaller \
--disable_pings \
--compress_fastq \
-c dna_r9.4.1_450bps_fast.cfg \
--num_callers 8 \
--gpu_runners_per_device 64 \
--chunks_per_runner 256 \
--device "cuda:0" \
--recursive \
-i flongle_fast5_pass \
-s testrun_fast_3.3.0
ONT Guppy basecalling software version 3.3.0+ef22818
config file: /home/miles/Downloads/software/guppy/3.3.0/ont-guppy/data/dna_r9.4.1_450bps_fast.cfg
model file: /home/miles/Downloads/software/guppy/3.3.0/ont-guppy/data/template_r9.4.1_450bps_fast.jsn
input path: flongle_fast5_pass
save path: testrun_fast_3.3.0
chunk size: 1000
chunks per runner: 256
records per file: 4000
fastq compression: ON
num basecallers: 8
gpu device: cuda:0
kernel path:
runners per device: 64
Found 105 fast5 files to process.
Init time: 722 ms
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
Caller time: 94953 ms, Samples called: 3970269916, samples/s: 4.1813e+07
Finishing up any open output files.
Basecalling completed successfully.
~/Downloads/software/guppy/3.3.3/ont-guppy/bin/guppy_basecaller \
--disable_pings \
--compress_fastq \
-c dna_r9.4.1_450bps_fast.cfg \
--num_callers 8 \
--gpu_runners_per_device 64 \
--chunks_per_runner 256 \
--device "cuda:0" \
--recursive \
-i flongle_fast5_pass \
-s testrun_fast_3.3.3
ONT Guppy basecalling software version 3.3.3+fa743a6
config file: /home/miles/Downloads/software/guppy/3.3.3/ont-guppy/data/dna_r9.4.1_450bps_fast.cfg
model file: /home/miles/Downloads/software/guppy/3.3.3/ont-guppy/data/template_r9.4.1_450bps_fast.jsn
input path: flongle_fast5_pass
save path: testrun_fast_3.3.3
chunk size: 1000
chunks per runner: 256
records per file: 4000
fastq compression: ON
num basecallers: 8
gpu device: cuda:0
kernel path:
runners per device: 64
Found 105 fast5 files to process.
Init time: 726 ms
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
Caller time: 95802 ms, Samples called: 3970269916, samples/s: 4.14425e+07
Finishing up any open output files.
Basecalling completed successfully.
~/Downloads/software/guppy/3.4.1/ont-guppy/bin/guppy_basecaller \
--disable_pings \
--compress_fastq \
-c dna_r9.4.1_450bps_fast.cfg \
--num_callers 8 \
--gpu_runners_per_device 64 \
--chunks_per_runner 256 \
--device "cuda:0" \
--recursive \
-i flongle_fast5_pass \
-s testrun_fast_3.4.1
ONT Guppy basecalling software version 3.4.1+ad4f8b9
config file: /home/miles/Downloads/software/guppy/3.4.1/ont-guppy/data/dna_r9.4.1_450bps_fast.cfg
model file: /home/miles/Downloads/software/guppy/3.4.1/ont-guppy/data/template_r9.4.1_450bps_fast.jsn
input path: flongle_fast5_pass
save path: testrun_fast_3.4.1
chunk size: 1000
chunks per runner: 256CUDA Version: 10.1
fastq compression: ON
num basecallers: 8
gpu device: cuda:0
kernel path:
runners per device: 64
Found 105 fast5 files to process.
Init time: 728 ms
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
Caller time: 90895 ms, Samples called: 3970269916, samples/s: 4.36797e+07
Finishing up any open output files.
Basecalling completed successfully.
~/Downloads/software/guppy/3.4.1/ont-guppy/bin/guppy_basecaller \
--disable_pings \
--compress_fastq \
-c dna_r9.4.1_450bps_fast.cfg \
--num_callers 8 \
--gpu_runners_per_device 64 \
--chunks_per_runner 256 \
--device "cuda:0" \
--recursive \
-i flongle_compressed \
-s testrun_fast_3.4.1
ONT Guppy basecalling software version 3.4.1+ad4f8b9
config file: /home/miles/Downloads/software/guppy/3.4.1/ont-guppy/data/dna_r9.4.1_450bps_fast.cfg
model file: /home/miles/Downloads/software/guppy/3.4.1/ont-guppy/data/template_r9.4.1_450bps_fast.jsn
input path: flongle_compressed
save path: testrun_fast_3.4.1
chunk size: 1000
chunks per runner: 256
records per file: 4000
fastq compression: ON
num basecallers: 8
gpu device: cuda:0
kernel path:
runners per device: 64
Found 105 fast5 files to process.
Init time: 725 ms
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
Caller time: 79913 ms, Samples called: 3970269916, samples/s: 4.96824e+07
Finishing up any open output files.
Basecalling completed successfully.
~/Downloads/software/guppy/3.4.3/ont-guppy/bin/guppy_basecaller \
--disable_pings \
--compress_fastq \
-c dna_r9.4.1_450bps_fast.cfg \
--num_callers 8 \
--gpu_runners_per_device 64 \
--chunks_per_runner 256 \
--device "cuda:0" \
--recursive \
-i flongle_fast5_pass \
-s testrun_fast_3.4.3_uncompressed
ONT Guppy basecalling software version 3.4.3+f4fc735
config file: /home/miles/Downloads/software/guppy/3.4.3/ont-guppy/data/dna_r9.4.1_450bps_fast.cfg
model file: /home/miles/Downloads/software/guppy/3.4.3/ont-guppy/data/template_r9.4.1_450bps_fast.jsn
input path: flongle_fast5_pass
save path: testrun_fast_3.4.3_uncompressed
chunk size: 1000
chunks per runner: 256
records per file: 4000
fastq compression: ON
num basecallers: 8
gpu device: cuda:0
kernel path:
runners per device: 64
Found 105 fast5 files to process.
Init time: 738 ms
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
Caller time: 270953 ms, Samples called: 3970269916, samples/s: 1.4653e+07
Finishing up any open output files.
Basecalling completed successfully.
ONT Guppy basecalling software version 3.4.3+f4fc735
config file: /home/miles/Downloads/software/guppy/3.4.3/ont-guppy/data/dna_r9.4.1_450bps_fast.cfg
model file: /home/miles/Downloads/software/guppy/3.4.3/ont-guppy/data/template_r9.4.1_450bps_fast.jsn
input path: flongle_fast5_pass
save path: testrun_fast_3.4.3_uncompressed
chunk size: 1000
chunks per runner: 256
records per file: 4000
fastq compression: ON
num basecallers: 8
gpu device: cuda:0
kernel path:
runners per device: 64
Found 105 fast5 files to process.
Init time: 705 ms
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
Caller time: 90674 ms, Samples called: 3970269916, samples/s: 4.37862e+07
Finishing up any open output files.
Basecalling completed successfully.
ONT Guppy basecalling software version 3.4.3+f4fc735
config file: /home/miles/Downloads/software/guppy/3.4.3/ont-guppy/data/dna_r9.4.1_450bps_fast.cfg
model file: /home/miles/Downloads/software/guppy/3.4.3/ont-guppy/data/template_r9.4.1_450bps_fast.jsn
input path: flongle_fast5_pass
save path: testrun_fast_3.4.3_uncompressed3
chunk size: 1000
chunks per runner: 256
records per file: 4000
fastq compression: ON
num basecallers: 8
gpu device: cuda:0
kernel path:
runners per device: 64
Found 105 fast5 files to process. Init time: 719 ms
0% 10 20 30 40 50 60 70 80 90 100% |----|----|----|----|----|----|----|----|----|----|
Caller time: 94516 ms, Samples called: 3970269916, samples/s: 4.20063e+07 Finishing up any open output files. Basecalling completed successfully.
~/Downloads/software/guppy/3.4.3/ont-guppy/bin/guppy_basecaller \
--disable_pings \
--compress_fastq \
-c dna_r9.4.1_450bps_fast.cfg \
--num_callers 8 \
--gpu_runners_per_device 64 \
--chunks_per_runner 256 \
--device "cuda:0" \
--recursive \
-i flongle_compressed \
-s testrun_fast_3.4.3
ONT Guppy basecalling software version 3.4.3+f4fc735
config file: /home/miles/Downloads/software/guppy/3.4.3/ont-guppy/data/dna_r9.4.1_450bps_fast.cfg
model file: /home/miles/Downloads/software/guppy/3.4.3/ont-guppy/data/template_r9.4.1_450bps_fast.jsn
input path: flongle_compressed
save path: testrun_fast_3.4.3
chunk size: 1000
chunks per runner: 256
records per file: 4000
fastq compression: ON
num basecallers: 8
gpu device: cuda:0
kernel path:
runners per device: 64
Found 105 fast5 files to process.
Init time: 721 ms
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
Caller time: 82877 ms, Samples called: 3970269916, samples/s: 4.79056e+07
Finishing up any open output files.
Basecalling completed successfully.
It's great to hear you've almost completed the transition. Following your past instructions and hard work, I've had great success running the MinION with fast basecalling for all the 16S analyses I do. Unfortunately, last week something strange happened (MinKNOW GUI no longer shows the different kits), so I've been forced to use my M1 MacBook... and the time differences are tremendous.