CellSAM is an advanced adaptation of the SAM, meticulously fine-tuned for the unique challenges posed by microscopy imaging. Microscopy images, especially those capturing cellular structures and activities, present intricate details that standard models might overlook. The inherent complexity, coupled with varying levels of noise, contrast, and sometimes even artifacts, necessitates a model with a heightened sense of focus and discernment, qualities epitomized by CellSAM
The genesis of CellSAM is rooted in the recognition that microscopy images, especially of cells, are replete with nuances. Cellular structures, organelles, and even intricate processes like mitosis, present patterns that are both intricate and crucial for accurate analysis. A model tailored for such images needs to be attuned not just to broad patterns but also to the minutiae, a task where the SAM excels
Whether it's imaging yeast cells, bacteria, or even multicellular organisms like zebrafish or C. elegans, CellSAM can be further fine-tuned or adapted. This extensibility ensures that researchers working across various biological domains can leverage the power of CellSAM for their specific needs
The Segment Anything Model (SAM) is an innovative image segmentation model developed by Meta AI, aiming to revolutionize object segmentation in images. The model is engineered to "cut out" or segment any object in any image with a simple click, showcasing a robust capability to generalize to unfamiliar objects and images without additional training.
SAM's architecture extends the realm of language prompting to visual prompting, a novel approach that facilitates zero-shot segmentation based on a prompt input. The prompts can vary, including a set of foreground/background points, free text, a box, or a mask, demonstrating the model's flexibility in handling diverse types of input for segmentation tasks.
The underlying architecture leverages the principles from foundation models that have significantly impacted natural language processing (NLP). By intertwining advanced machine learning techniques with practical utility, SAM's architecture manifests as a blend of innovative design and functional prowess, aiming to significantly advance the field of image segmentation and analysis in computer vision.
SAM's architecture comprises of:
- Prompting Mechanism: Allows for versatile input prompts to guide the segmentation process.
- Zero-shot Generalization: Enables the model to adapt to unfamiliar objects and images without re-training.
- Visual Prompting Extension: An extension of language prompting techniques to the visual domain, enriching the model's segmentation capability.
The Segment Anything Model, with its groundbreaking architecture, not only stands as a testament to the advancements in image segmentation but also as a substantial contribution to the broader field of computer vision, paving the way for more nuanced and accurate image analysis.
Cell segmentation is a cornerstone in the expansive field of biological and medical research. It's the precise art and science of identifying and delineating individual cells in microscopy images, opening up a realm of possibilities for analyzing cellular structures, behaviors, and interactions. From understanding cell morphology to unraveling the effects of drugs on cells, the domain of cell segmentation is as vast as it is vital.
Our endeavor is to take the robust Segment Anything Model (SAM) and fine-tune it to excel in the domain of microscopy image segmentation, going beyond its original training on color images. SAM, with its promptable segmentation system, provides a solid foundation to build upon. Our project aims to adapt SAM's architecture and pre-trained weights, making it adept at navigating the nuanced world of microscopy images.
Leveraging SAM’s pre-trained weights and architecture, we venture into the domain of microscopy images. The process of fine-tuning is not merely a technical adjustment but an enriching interaction between the model and the microscopic realm. It’s about nurturing SAM, guiding its evolution to develop a refined understanding of cellular structures, morphologies, and interactions amidst the complex tapestry of microscopy images.
Through the lens of domain-specific datasets, we steer SAM through the currents of fine-tuning. Each adjustment, each iteration brings SAM closer to accurately segmenting cells, understanding the subtle intricacies of cellular morphologies, and decoding the complex narrative told by clusters of cells.
The adaptation of SAM is more than just a technical endeavor; it's about ushering in a new era of cellular analysis. With SAM fine-tuned for microscopy images, we unlock a treasure trove of insights. It’s not merely about segmenting cells; it’s about enabling a deeper understanding, a clearer visualization of the cellular cosmos.
Our fine-tuned SAM model is poised to be a valuable asset for researchers navigating the complex waters of cellular biology, oncology, and drug discovery. It's about providing a refined tool, a companion in the quest to unravel the intricacies of cellular structures and behaviors, fueling the engines of discovery in life sciences.
This project embodies the convergence of advanced machine learning and practical utility, aiming to significantly contribute to the broader field of bioimage analysis, and propelling the wheel of discovery in the life sciences. Thank you for the clarification. Here's a more refined overview considering the embryonic stages of C. elegans you've mentioned:
The embryogenesis of Caenorhabditis elegans (C. elegans) is a highly orchestrated and deterministic process. The transparent nature of C. elegans, coupled with its invariant cell lineage, makes it a particularly attractive model organism for studying the intricacies of embryonic development.
During embryogenesis, C. elegans undergoes a series of well-defined cellular divisions, beginning with a single fertilized egg and culminating in a young worm with all its somatic cells. The stages you mentioned, from 2 to 360 cells, represent the early to mid-phases of this embryonic development.
-
2-cell stage: Shortly after fertilization, the zygote undergoes its first asymmetric division to produce two distinct daughter cells: the larger anterior cell (AB) and the smaller posterior cell (P1). Each of these cells will give rise to specific tissues in the mature worm.
-
Subsequent stages: As development proceeds, these initial cells divide further, following a consistent and well-documented pattern. By the 360-cell stage, a significant portion of the cell lineage has been established, with many of the cells destined to become specific tissues, organs, or structures in the mature worm.
-
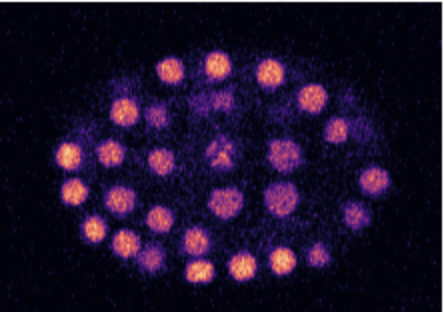
Microscopy Image: These images capture the embryonic stages of C. elegans, allowing researchers to observe the spatial arrangement, morphology, and interactions of cells during the early phases of development.
-
Segmentation Map: At the embryonic stage, individual cells or groups of cells undergo rapid divisions. The segmentation maps help delineate each cell, making it easier to track cellular lineage and fate as the embryo develops.

-
Centroid of the Cell: Visualizing the centroids during embryogenesis provides insights into how cells move, orient themselves, and interact during the development process. Tracking these centroids can help researchers understand the choreography of cellular divisions and migrations.

-
Centroid of the Cell: Visualizing the centroids during embryogenesis provides insights into how cells move, orient themselves, and interact during the development process. Tracking these centroids can help researchers understand the choreography of cellular divisions and migrations.
Given the deterministic nature of C. elegans development, this dataset offers invaluable insights into the cellular and molecular mechanisms driving embryogenesis. It can be used to investigate how genes regulate developmental processes, understand the biophysical properties of developing tissues, and study the coordination and timing of cellular events during embryonic development.
The SegmentationDataset class is intricately designed to address the challenges of image segmentation, especially for datasets like the nucleus dataset. During its initialization, the class reads from a specified CSV file to extract file IDs and establishes paths to retrieve the corresponding images and segmentation masks. One of its standout features is the ability to incorporate bounding box shifts, which can be fine-tuned during initialization.
When the dataset is queried for an entry, it fetches the associated image and mask, standardizing their sizes to a uniform 1024x1024 resolution. Once resized, the image is normalized to have values between 0 and 1. Given that the images in the dataset can have varying numbers of cells, there's an inherent challenge in consistently selecting labels for segmentation. This variability leads to the problem of generating different numbers of bounding boxes for each image, causing inconsistencies during batch loading in data loaders. To navigate this challenge, the class transforms the mask into a binary format by randomly selecting a single cell label. This approach ensures a uniform output structure, allowing for easier batch processing. While using a collate function could address this issue, it requires a substantial amount of GPU resources. Therefore, to make the process more efficient and resource-friendly, the dataset class focuses on segmenting only one cell and provides its bounding box for segmentation.
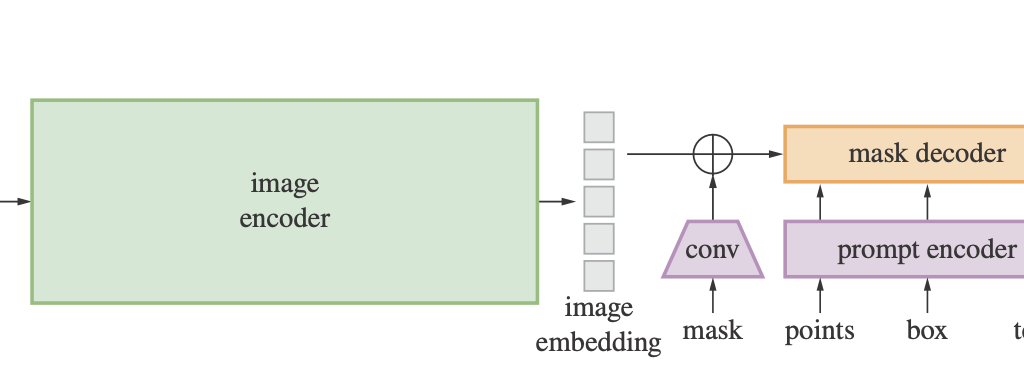
CellSAM stands as a testament to the harmony achieved between an image encoder, mask decoder, and a prompt encoder, all working in tandem to perfect the art of image segmentation. Upon its initialization, while the fundamental components are anchored in place, the weights of the prompt and image encoders are frozen. This strategic move ensures that these modules retain their pre-trained prowess and remain invariant during the training process.
In the forward pass, the journey of the input image begins with the image encoder, culminating in an embedding. This embedding, when paired with the sparse and dense embeddings from the gradient-free prompt encoder, flows into the mask decoder. The end product is a low-resolution mask, which is then meticulously rescaled to resonate with the original resolution of the input image. Such an architectural marvel not only leverages the combined strengths of image and prompt information but also sets new standards in mask prediction accuracy, making it indispensable for complex segmentation tasks.
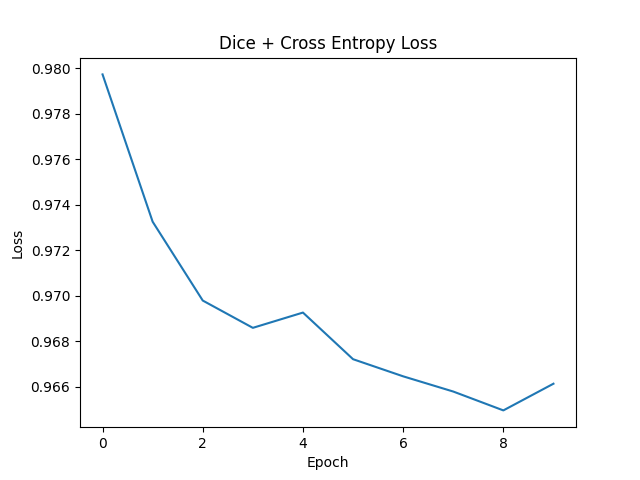
In the provided code, two prominent loss functions are combined to optimize a segmentation task: the Dice Loss and the Binary Cross-Entropy with Logits Loss (BCEWithLogitsLoss). Let's delve into each of these losses to comprehend their working mechanisms and significance.
Dice Loss, often referred to as the Sørensen–Dice coefficient, is a renowned loss function primarily employed for binary and multi-class segmentation. Its primary role is to gauge the similarity between two samples. Mathematically, it's articulated as:
For a scenario of perfect overlap, the Dice Loss evaluates to 0, while for no overlap, it amounts to 1.
In the provided snippet, the Dice Loss from MONAI is instantiated with the subsequent configurations:
sigmoid=True: Enforces the application of the sigmoid activation function on the predicted output, ensuring its range is confined between 0 and 1.squared_pred=True: Elevates the predictions post the sigmoid activation, an approach that can occasionally aid in streamlining the training by casting emphasis on false positives and false negatives.reduction="mean": Computes the mean Dice Loss across all batch samples.
This loss amalgamates the sigmoid activation function and the binary cross-entropy loss into a unified class, offering enhanced numerical stability compared to their individual applications. Its mathematical representation is:
Here, y symbolizes the ground truth, and p denotes the predicted probability.
Within the code:
reduction="mean": Calculates the mean binary cross-entropy loss over the entire batch.
The essence of the code lies in the fusion of the aforementioned losses:
Such a union capitalizes on the strengths of both loss functions. While the Dice Loss is centered around the overlap between the predicted segments and the ground truth, BCEWithLogitsLoss penalizes individual pixel classification. This synergy encourages the model to holistically focus on both pixel-level classification and segment overlap, offering a more rounded training impetus.
In a nutshell, for segmentation tasks, the blend of Dice Loss and BCEWithLogitsLoss can potentially usher in superior segmentation outcomes, given that the model is concurrently guided by both pixel-level classifications and segment overlap considerations.
The primary focus of our GSoC project was to advance the capabilities of segmentation models for microscopy images. Below are the summarized results and conclusions based on the efforts made:
-
Design and Purpose: This model has been architecturally tailored to operate based on bounding box prompts, providing a unique approach to segmentation tasks.
- Design and Purpose: In contrast to the bounding box model, this variant operates using point prompts, offering another dimension of flexibility in segmentation.
-
Loss Function Optimization:
-
Multi-GPU Support:
- Recognizing the need for high computational power and efficiency, multi-GPU support was seamlessly integrated.
- This feature paves the way for simultaneous training on multiple GPUs, translating to swifter training cycles and optimal resource utilization.
-
Tutorial Development:
- A thorough tutorial was curated to facilitate users in employing pretrained weights for their segmentation endeavors.
- This initiative ensures the model's accessibility and usability, even for individuals who might be novices in the domain.
-
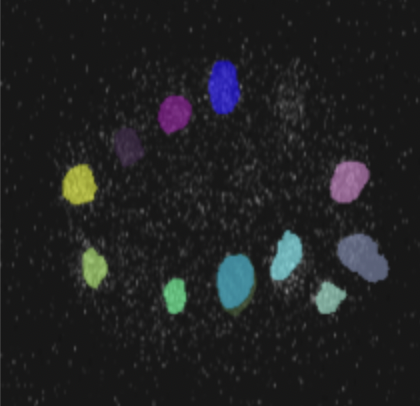
Segmentation Proficiency:
- The showcased image stands as a testament to the model's segmentation prowess.
- The distinct coloration for various segments underscores the model's accuracy and precision in discerning different entities within a microscopy image.
In wrapping up, the strides made in this project have significantly bolstered the segmentation capabilities for microscopy images. The models developed, equipped with the aforementioned features, are poised to make a marked difference in the domain of image segmentation.
- CLIP Model Integration: Incorporating the CLIP model for a combined visual and linguistic approach promises enhanced cell lineage analysis.
- Expansion to Other Organisms: Beyond C. elegans, the model harbors the potential for adaptability to other model organisms and live cell segmentation.
Plans are underway to host the models on HuggingFace Spaces, offering users easy access and the ability to seamlessly segment microscopy images. Furthermore, considerations are being made to render the model as a dedicated tool for cell segmentation, ensuring broader usability for the scientific community.
As I reflect upon the journey of this GSoC project, I find myself overwhelmed with gratitude. A special thanks to my mentors, Bradley, Mainak, and Mayukh Deb, whose unwavering support, insightful guidance, and patient mentorship were instrumental in turning our vision into a successful reality. Their expertise and encouragement fueled my passion and determination throughout the project.
I'd also like to extend my heartfelt appreciation to the OpenWorm Foundation for providing me with this incredible opportunity and to the Google Summer of Code for fostering such a nurturing environment for budding developers like myself.
Thank you all for believing in me and for being a part of this transformative experience.
Sources: