- Name: Haroune Hassine
- Email: thessalonikaathena@outlook.com
- Home Country: Tunisia
- GitHub: waterflow80
- Organization: Global Alliance for Genomics and Health (GA4GH)
- Project Name: Implementation of the Sequence collection specification using the EVA contig alias backend
- Specification being Implemented: Seqcol specification version 0.1.0
- Mentors:
- Timothee Cezard
- April Shen
- Sundar Venkataraman
- Project Link: github.com/EBIvariation/eva-seqcol
- Project Proposal: Gsoc 23' Proposal
- Deliverables: GSoC'23 Deliverables - GA4GH
The genome of an organism consists of a set of chromosomes (or contigs). Genomic scientists often rely on using a reference genome to define a baseline of knowledge against which our understanding of biological systems, phenotypes and variation are based upon. A majority of these genomes are served by central providers such as INSDC (e.g. NCBI, ENA), Ensembl or UCSC and if they most of the time agree on the sequence being used they often differ on the naming of these sequences. Thus a standardized gene naming is crucial for effective communication about genes.
In this context, a service named the Contig Alias service has been developed by the EVA for the purpose of providing a translation between different sequence naming conventions.
In the same context, the GA4GH has created a standard named Sequence Collection (SeqCol) that aims mainly at providing a protocol for uniquely identifying the content of a set of sequences and a comparison function that assess the compatibility of 2 provided sequence collections.
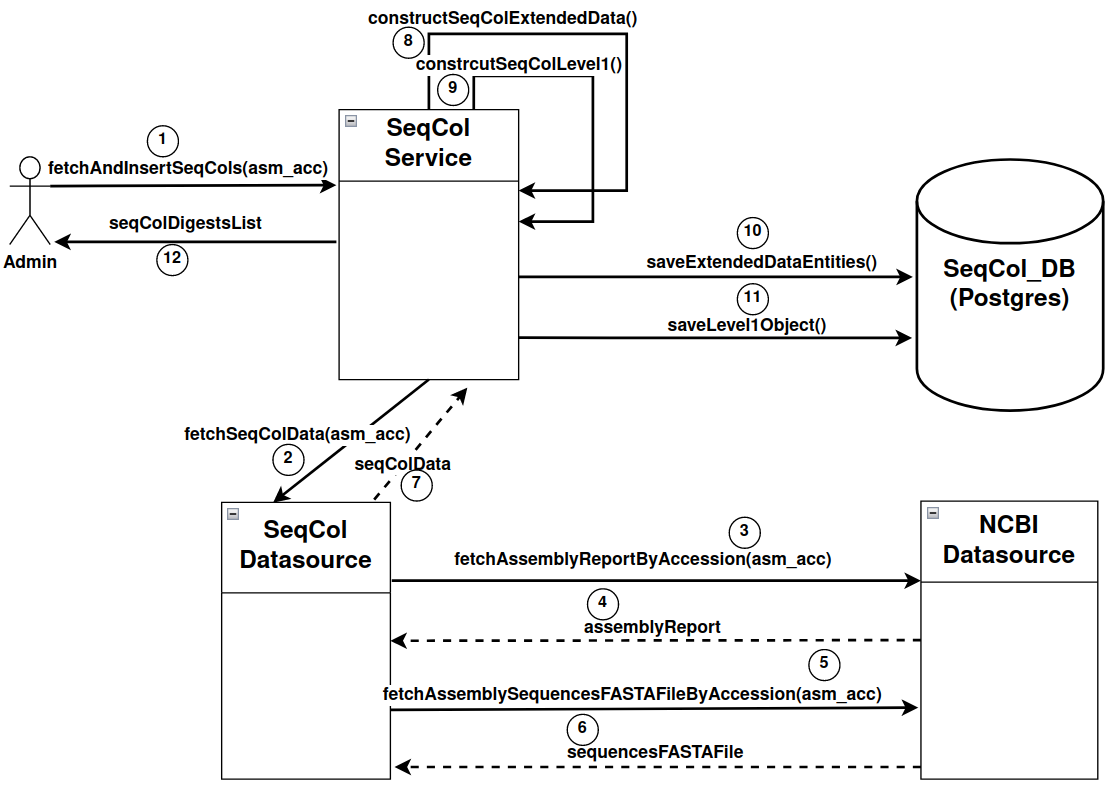
- Evaluation of the contig alias database and data model
- Create the databases and Java model to represent a reference genome as sequence collection data (link)
- Create a mechanism to serialise and digest a sequence collection object.
- Define the main endpoint that will resolve the sequence collection object in json from the digest (link)
- Create a comparison function and the endpoint associated (link)
 For more details about the sequence digests computation, see seqcol.readthedocs.io
For more details about the sequence digests computation, see seqcol.readthedocs.io
In order to make an efficient data model for the seqCol porject, we've chosen the disk (db) space and the processing time as the main criteria of choosing this model. In this process, we've made some evaluations of different data-models like:
- Evaluation of the contig-alias backend to determine whether it is useful to use it's data-model for our API. Eventually we agreed that this data model is not the best option for us. More details about the evaluation of this model can be found here.
- Evaluation of a recursive data-model that uses only one database table to store all different levels of seqCol objects. This model has also been rejected. More details about the evaluation of this model can be found here.
- Evaluation of a recursive data-model, which is the model we've chosen to use, that uses two database tables to store the seqCol objects' data in different levels. More details about the evaluation of this model can be found here. The design of this model looks like this:

-
sequence_collections_l1: stores the seqCol level 1 objects. Example:
digest (level 0) naming_convention seq_col_level1object rkTW1yZ0e22IN8K-0frqoGOMT8dynNyE UCSC {"names": "rIeQU2I79mbAFQwiV_kf6E8OUIEWq5h9", "lengths": "Ms_ixPgQMJaM54dVntLWeovXSO7ljvZh", "sequences": "dda3Kzi1Wkm2A8I99WietU1R8J4PL-D6", "md5-sequences": "_6iaYtcWw4TZaowlL7_64Wu9mbHpDUw4", "sorted-name-length-pairs": "qCETfSy_Ygmk0qtUJwI8V6SdsYX_AC53"}3mTg0tAA3PS-R1TzelLVWJ2ilUzoWfVq GENBANK {"names": "mfxUkK3J5y7BGVW7hJWcJ3erxuaMX6xm", "lengths": "Ms_ixPgQMJaM54dVntLWeovXSO7ljvZh","sequences": "dda3Kzi1Wkm2A8I99WietU1R8J4PL-D6", "md5-sequences": "_6iaYtcWw4TZaowlL7_64Wu9mbHpDUw4", "sorted-name-length-pairs": "QFuKs5Hh8uQwwUtnRxIf8W3zeJoFOp8Z"} -
seqcol_extended_data: stores the seqCol extended (exploded) level 2 seqCol data. Each entry has a digest that corresponds to one of the seqCol level 1 attribute's values. Example:
digest extended_seq_col_data dda3Kzi1Wkm2A8I99WietU1R8J4PL-D6 {"object": ["SQ.lZyxiD_ByprhOUzrR1o1bq0ezO_1gkrn", "SQ.vw8jTiV5SAPDH4TEIZhNGylzNsQM4NC9", "SQ.A_i2Id0FjBI-tQyU4ZaCEdxRzQheDevn", "SQ.QXSUMoZW_SSsCCN9_wc-xmubKQSOn3Qb", "SQ.UN_b-wij0EtsgFqQ2xNsbXs_GYQQIbeQ", "SQ.z-qJgWoacRBV77zcMgZN9E_utrdzmQsH", "SQ.9wkqGXgK6bvM0gcjBiTDk9tAaqOZojlR", "SQ.K8ln7Ygob_lcVjNh-C8kUydzZjRt3UDf", "SQ.hb1scjdCWL89PtAkR0AVH9-dNH5R0FsN", "SQ.DKiPmNQT_aUFndwpRiUbgkRj4DPHgGjd", "SQ.RwKcMXVadHZub1qL0Y5c1gmNU1_vHFme", "SQ.1sw7ZtgO9JRb1kUEuhVz1wBix5_8Opci", "SQ.V7DQqMKG7bcyxiMZK9wNjkK-udR7hrad", "SQ.R8nT1N2qQFMc_uVMQUVMw-D2GcVmb5v6", "SQ.DPa_ORXLkGyyCbW9SWeqePfortM-Vdlm", "SQ.koyLEKoDOQtGHjb4r0m3o2SXxI09Z_sI"]}_6iaYtcWw4TZaowlL7_64Wu9mbHpDUw4 ... rIeQU2I79mbAFQwiV_kf6E8OUIEWq5h9 ... Ms_ixPgQMJaM54dVntLWeovXSO7ljvZh ... qCETfSy_Ygmk0qtUJwI8V6SdsYX_AC53 ... mfxUkK3J5y7BGVW7hJWcJ3erxuaMX6xm ... QFuKs5Hh8uQwwUtnRxIf8W3zeJoFOp8Z ...
The digests in bold indicates how the data is handled recursively, pretty much, between the two tables.
A detailed evaluation of all different proposed data-models can be found here: Deliverable 1.
Note: the seqCol service is currently deployed on server 45.88.81.158, under port 8081
PUT - SERVER_IP:PORT/eva/webservices/seqcol/admin/seqcols/{asm_accession}GET - SERVER_IP:PORT/eva/webservices/seqcol/collection/{seqCol_digest}?level={level}GET - SERVER_IP:PORT/eva/webservices/seqcol/comparison/{seqColA_digest}/{seqColB_digest}POST - SERVER_IP:PORT/eva/webservices/seqcol/comparison/{seqColA_digest}; body = {level 2 JSON representation of another seqCol}GET - SERVER_IP:PORT/eva/webservices/seqcol/collection/service-info
For a more detailed, user-friendly documentation of the API's endpoints, please visit the seqCol's swagger page
PUT - SERVER_IP:PORT/eva/webservices/seqcol/admin/seqcols/{asm_accession}- Description: Ingest all possible (all possible naming conventions) seqCol objects in the database for the given assembly accession and return the list of digests of the inserted seqCol objects
- Permission: Authenticated endpoint (requires admin privileges)
- Example: PUT - http://45.88.81.158:8081/eva/webservices/seqcol/admin/seqcols/GCA_000146045.2
GET - SERVER_IP:PORT/eva/webservices/seqcol/collection/{seqCol_digest}?level={level}- Description: Retrieve the seqCol object that has the given seqCol_digest in the given level representation (level should be either 1 or 2, default=2)
- Permission: Public endpoint
- Example: GET - http://45.88.81.158:8081/eva/webservices/seqcol/collection/3mTg0tAA3PS-R1TzelLVWJ2ilUzoWfVq
GET - SERVER_IP:PORT/eva/webservices/seqcol/comparison/{seqColA_digest}/{seqColB_digest}- Description: Compare two seqCol objects given their level 0 digests
- Permission: Public endpoint
- Example: GET - http://45.88.81.158:8081/eva/webservices/seqcol/comparison/rkTW1yZ0e22IN8K-0frqoGOMT8dynNyE/3mTg0tAA3PS-R1TzelLVWJ2ilUzoWfVq
POST - SERVER_IP:PORT/eva/webservices/seqcol/comparison/{seqColA_digest}; body = {level 2 JSON representation of another seqCol}- Description: Compare two seqCol objects given the first's level 0 digest, and the other's level 2 JSON representation
- Permission: Public endpoint
- Example: POST - http://45.88.81.158:8081/eva/webservices/seqcol/comparison/rkTW1yZ0e22IN8K-0frqoGOMT8dynNyE, Body =
{ "sequences": [ "SQ.lZyxiD_ByprhOUzrR1o1bq0ezO_1gkrn", "SQ.vw8jTiV5SAPDH4TEIZhNGylzNsQM4NC9", "SQ.A_i2Id0FjBI-tQyU4ZaCEdxRzQheDevn" ], "names": [ "I", "II", "III" ], "lengths": [ "230218", "813184", "316620" ] }GET - SERVER_IP:PORT/eva/webservices/seqcol/collection/service-info- Description: Get the service info page of the API. For more details, please see ga4gh/seqcol-spec/issues/39
- Permisssion: Public endpoint
- Example: http://45.88.81.158:8081/eva/webservices/seqcol
This web service has some authenticated endpoints. The current approach to secure them is to provide the credentials in the src/main/resources/application.properties file at compilation time, using maven profiles.
The application also requires to be connected to an external database (PostgreSQL by default) to function. The credentials for this database need to be provided at compilation time using the same maven profiles.
You can edit the maven profiles values in pom.xml by locating the below section and changing the values manually or by setting environment variables. Alternatively, you can make the changes directly on the application.properties file.
Use <ftp.proxy.host> and <ftp.proxy.port> to configure proxy settings for accessing FTP servers (such as NCBI's). Set them to null and 0 to prevent overriding default the proxy configuration.
Set a boolean flag using <contig-alias.scaffolds-enabled> to enable or disable parsing and storing of scaffolds in the database.
<profiles>
<profile>
<id>seqcol</id>
<properties>
<spring.profiles.active>seqcol</spring.profiles.active>
<seqcol.db-url>jdbc:postgresql://${env.SERVER_IP}:${env.POSTGRES_PORT}/seqcol_db</seqcol.db-url>
<seqcol.db-username>${env.POSTGRES_USER}</seqcol.db-username>
<seqcol.db-password>${env.POSTGRES_PASS}</seqcol.db-password>
<seqcol.ddl-behaviour>${env.DDL_BEHAVIOUR}</seqcol.ddl-behaviour>
<seqcol.admin-user>${env.ADMIN_USER}</seqcol.admin-user>
<seqcol.admin-password>${env.ADMIN_PASSWORD}</seqcol.admin-password>
<ftp.proxy.host>${optional default=null}</ftp.proxy.host>
<ftp.proxy.port>${optional default=0}</ftp.proxy.port>
<contig-alias.scaffolds-enabled>${optional default=false}</contig-alias.scaffolds-enabled>
</properties>
<activation>
<activeByDefault>true</activeByDefault>
</activation>
</profile>
</profiles>
Once that's done, you can trigger the variable replacement with the -P option in maven. Example: mvn clean install -Pseqcol to compile the service including tests or mvn clean install -Pseqcol -DskipTests to ignore tests.
You can then run: mvn spring-boot:run to run the service.
The current version of our API is currently deployed in an embedded tomcat server at 45.88.81.158 under port 8081. Briefly, the deployment steps are as follows:
- Connect to server
45.88.81.158(deployment server) via SSH - Clone the seqCol project's repo on the server (under /opt/eva-seqcol for example)
- Update the configuration files such as
pom.xmland/orapplication.propertieswith the correct information/credentials - Run: /opt/eva-seqcol$ nohup bash -c "git pull && mvn clean install -DskipTests -f local-pom.xml && export TOMCAT_PASS=`cat ~/.seqcol-tomcat-pass` && sleep 5 && export WAR_FILE=`ls -1 target/*.war|head -1` && java -jar $WAR_FILE" 1>> eva-seqcol.log 2>&1 &
- Log messages are in eva-seqcol.log file, you can see them live running the command /opt/eva-seqcol$ tail -f eva-seqcol.log
- If you want to stop the running version and pull the latest version, you can run: /opt/eva-seqcol$ kill `ps -ef | grep -E '.[e]va-seqcol..war' | awk '{print $2}'`, and repeat steps from 4.
- Issue #105: Sequence collection data models using contig alias
- Issue #106: Sequence collection data models without using contig alias
- Issue #107: Add function to download the fasta file when retrieving the genome from FTP
- Issue #108: Some assemblies in the NCBI database does not have a fna/fasta files
- Issue #111: Evaluation of the Contig-alias's data model
- PR #110: Seqcol: Retrieve the FASTA file from the reomte repository.
- PR #112: With contig alias (Data Model Proposition)
- PR #113: Recursive approach (Data Model Proposition)
- PR #114: Recursive approach v2 (Data Model Proposition)
- PR #115: Recursive approach v3 (Data Model Proposition)
- PR #116: Relational data model (Data Model Proposition)
- PR #117: Simple intuitive approach (Data Model Proposition)
- PR #1: Created the spring boot project with the necessary dependencies
- PR #2: Implemented the process of downloading the assembly sequences fasta file
- PR #3: Changed the checksum calculation design and renamed some classes plus slight changes
- Issue #4: Refget checksum calculation (ga4gh ) not working
- Issue #5: Confusing sequence (chromosome) identifiers in the FASTA files
- PR #6: Composite primary key
- PR #7: Ingest seqcol process
- PR #8: Create maven.yml - Github action
- PR #9: Data model refactor
- PR #10: Full ingest of seqcol
- PR #11: Seqcol initial controller
- Issue #12: Some naming conventions are missing in some assembly reports
- PR #14: Add new attribute to model
- PR #16: Seqcol comparison api
- Issue #18: Method getSeqColByDigestAndLevel unexpectedly throws org.hibernate.LazyInitializationException...
- PR #19: fixed the digest calculation algorithm
- PR #20: Seqcol comparison api generic version - using low level reflection libraries
- PR #21: added the unbalanced-duplicate condition to the comparison api
- PR #22: fetch and insert all possible seqCol objects from one assembly read
- Issue #23: Adding flexibility for additional attributes in comparison endpoint
- PR #24: Test suite enhancements - SeqColWriter & AssemblyDataGenerator
- Issue #26: There's still a use of the language level reflections in the merged code
- Issue #27: Common elements count algorithm is not correct - Comparison api
- PR #28: fix common elements count algorithm - Comparison API
- PR #29: Change json attribute name - md5-sequences
- PR #30: added the sorted-name-length-pair attribute to the model
- PR #31: Some code clean up and enhancements
- Issue #32: Using the wrong algorithm to digest seqCol Level 2 into Level 1 seqCol object
- PR #33: Generic comparison function for both GET and POST requests
- Issue #34: Wrong algorihtm for a-and-b-same-order in comparison function
- PR #36: Digest algorithm correction - level 2 to level 1 digest calculation
- Issue #37: Declare implementation capability in service-info endpoint
- Issue #38: Create swagger page
- PR #40: fixed the a-and-b-same-order algorithm
- PR #41: added a basic authentication for admin endpoints
- PR #42: Swagger documentation
- PR #43: Implemented the service-info endpoint
- PR #44: basic integration tests for the API's endpoints
- PR #46: Fixed the digits verification for seqCol arrays to be digested - ingestion endpoint
- PR #47: fixed the seqCol retrieval's default level
- Spring Boot v2.7.13
- PostgreSQL Database v15.2
- Swagger v3 (springdoc-openapi implementation)
- Git and GitHub Actions
- seqCol, seqcol-spec, specification (Specification's details and docs)
- GA4GH refget API meetings (Minutes for the refget API meetings)
- Python implementation (A python implementation of the sequence collection specification)
- CRAM Reference Registry
Google Summer of Code was one the best programs that I've worked in so far. During this program, I had the opportunity to work and collaborate with highly experienced people wtih whom I gained, not only a lot of technical skills in Java, Spring Boot, Git, GitHub etc, but also a lot of project management skills that allowed me to submit deliverables in time. So I can list my gained experience in GSoC '23 as follows:
- Enhanced my programming skills mainly in Java and Spring Boot
- Enhanced my software and database design capabilities
- Enhanced my skills in Git, GitHub actions and open source contribution in general
- Enhanced my communication skills that allowed me to better reveal my ideas and ask the correct questions
- Got involved with a wonderful community full of passion and energy
Finally, I would like to give special thanks to the people who helped making this project successful, which are my mentors: Timothee Cezard, April Shen and of course Sundar Venkataraman . They were so professional and supportive and I was really lucky to have them as my mentors.