Having fun with Ramnath Vaidyanathan's new package rblocks!
Last active
August 29, 2015 13:57
-
-
Save woobe/9624370 to your computer and use it in GitHub Desktop.
[rblocks]: Experiments
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
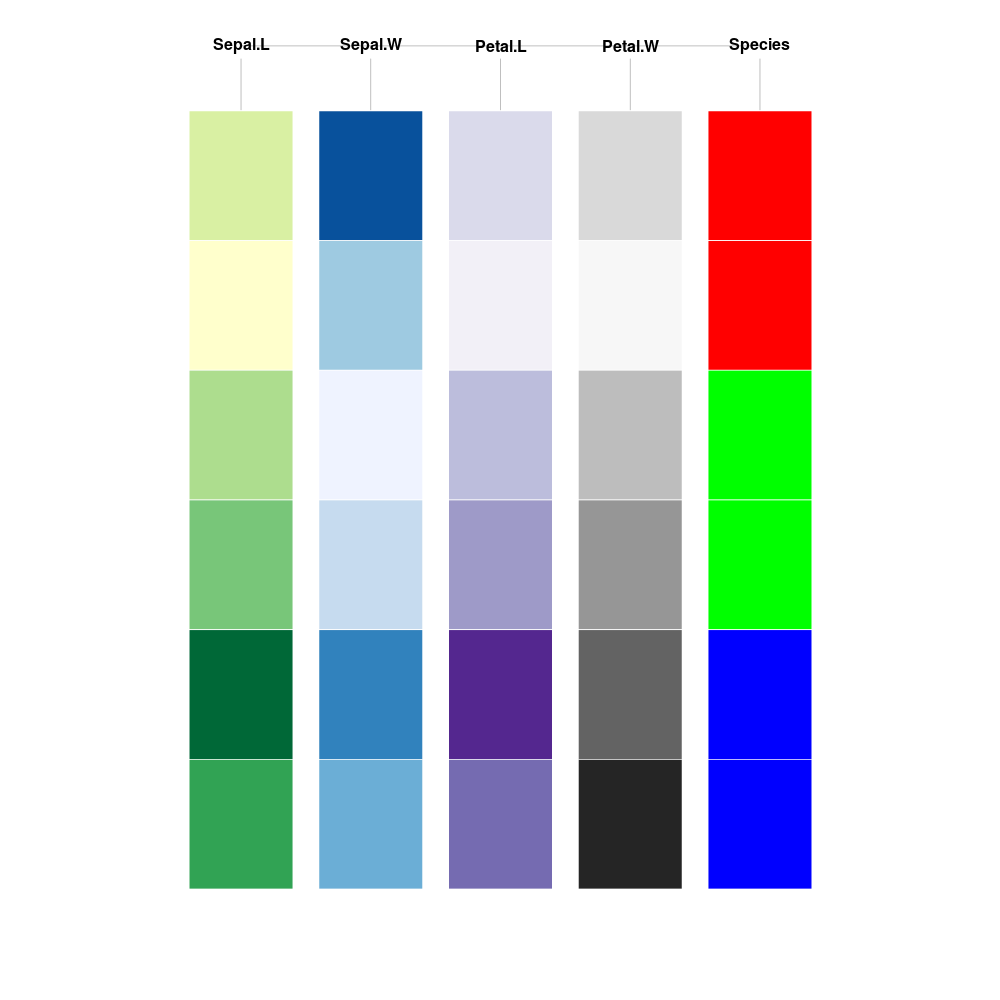
| ## Load packages (see https://github.com/ramnathv/rblocks) | |
| library(caret) | |
| library(rblocks) | |
| library(RColorBrewer) | |
| ## Sampling with caret (to get exactly two samples from each species) | |
| set.seed(1234) | |
| row_samp <- createDataPartition(iris$Species, p = 0.021, list = F) | |
| ## Make block | |
| iris_samp <- iris[row_samp,] | |
| iris_block <- make_block(iris_samp) | |
| ## Shorten column names for better display | |
| colnames(iris_block) <- c("Sepal.L", "Sepal.W", "Petal.L", "Petal.W", "Species") | |
| ## Update colours for Species (ref: http://en.wikipedia.org/wiki/File:Anderson%27s_Iris_data_set.png) | |
| iris_block[which(iris_samp$Species == "setosa"),]$Species <- "red" | |
| iris_block[which(iris_samp$Species == "versicolor"),]$Species <- "green" | |
| iris_block[which(iris_samp$Species == "virginica"),]$Species <- "blue" | |
| ## Update colours for other variables | |
| iris_block[order(iris_samp[,1]), 1] <- colorRampPalette(brewer.pal(6,"YlGn"))(length(row_samp)) | |
| iris_block[order(iris_samp[,2]), 2] <- colorRampPalette(brewer.pal(6,"Blues"))(length(row_samp)) | |
| iris_block[order(iris_samp[,3]), 3] <- colorRampPalette(brewer.pal(6,"Purples"))(length(row_samp)) | |
| iris_block[order(iris_samp[,4]), 4] <- colorRampPalette(brewer.pal(6,"Greys"))(length(row_samp)) | |
| ## Plot | |
| iris_block |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| ## Load packages | |
| library(rblocks) | |
| library(RColorBrewer) | |
| ## ============================================================================= | |
| ## rBlocks #1 - AirPassengers with 'YlOrRd' palette | |
| ## ============================================================================= | |
| ## Using the classic AirPassengers dataset | |
| mat_air <- matrix(AirPassengers) | |
| mat_value <- matrix(mat_air, ncol = 12, byrow = TRUE) | |
| ## Assign colours to numerical values | |
| colour_value <- matrix(NA, nrow(mat_air)) | |
| colour_value[order(mat_air),] <- colorRampPalette(brewer.pal(9,"YlOrRd"))(nrow(mat_air)) | |
| colour_value <- matrix(colour_value, ncol = 12, byrow = TRUE) | |
| ## Assign colours to years | |
| mat_year <- matrix(1949:1960) | |
| colour_year <- colorRampPalette(brewer.pal(6,"Greys"))(nrow(mat_year)) | |
| ## Create df | |
| df_air <- data.frame(mat_year, mat_value) | |
| colnames(df_air) <- c("Year", | |
| "Jan", "Feb", "Mar", "Apr", "May", "Jun", | |
| "Jul", "Aug", "Sep", "Oct", "Nov", "Dec") | |
| ## Make block | |
| block_air <- make_block(df_air) | |
| ## Update Colours | |
| block_air[, 1] <- as.character(colour_year) | |
| block_air[, -1] <- as.character(colour_value) | |
| ## Plot | |
| block_air | |
| ## ============================================================================= | |
| ## rBlocks #2 - AirPassengers with funky RSkittleBrewer | |
| ## ============================================================================= | |
| ## Using the RSkittleBrewer | |
| if (!require(devtools)) install.packages("devtools") | |
| if (!require(RSkittleBrewer)) devtools::install_github('RSkittleBrewer', 'alyssafrazee') | |
| ## Assign Skittle (original) colours to numerical values | |
| colour_skittle <- RSkittleBrewer('original') | |
| colour_value <- matrix(NA, nrow(mat_air)) | |
| colour_value[order(mat_air, decreasing = T),] <- colorRampPalette(colour_skittle)(nrow(mat_air)) | |
| colour_value <- matrix(colour_value, ncol = 12, byrow = TRUE) | |
| ## Make block | |
| block_air_skittle <- make_block(df_air) | |
| ## Update Colours | |
| block_air_skittle[, 1] <- as.character(colour_year) | |
| block_air_skittle[, -1] <- as.character(colour_value) | |
| ## Plot | |
| block_air_skittle |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| ## Load packages | |
| library(rblocks) | |
| library(RColorBrewer) | |
| ## ============================================================================= | |
| ## Core Parameters | |
| ## ============================================================================= | |
| side <- 50 # side - side of the game of life arena (matrix) | |
| steps <- 170 # steps - number of animation steps | |
| skips <- 20 # skips - initial frames to be dropped (burn out) | |
| set.seed(3) # Set seed for reproducible results | |
| ## ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ | |
| ## Original Game of Life Arena Codes | |
| ## http://www.r-bloggers.com/fast-conways-game-of-life-in-r/ | |
| ## ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ | |
| # the sideXside matrix, filled up with binomially | |
| # distributed individuals | |
| X <- matrix(nrow=side, ncol=side) | |
| X[] <- rbinom(side^2,1,0.4) | |
| # array that stores all of the simulation steps | |
| # (so that it can be exported as a gif) | |
| storage <- array(0, c(side, side, steps)) | |
| # the simulation | |
| for (i in 1:steps) | |
| { | |
| # make the shifted copies of the original array | |
| allW = cbind( rep(0,side) , X[,-side] ) | |
| allNW = rbind(rep(0,side),cbind(rep(0,side-1),X[-side,-side])) | |
| allN = rbind(rep(0,side),X[-side,]) | |
| allNE = rbind(rep(0,side),cbind(X[-side,-1],rep(0,side-1))) | |
| allE = cbind(X[,-1],rep(0,side)) | |
| allSE = rbind(cbind(X[-1,-1],rep(0,side-1)),rep(0,side)) | |
| allS = rbind(X[-1,],rep(0,side)) | |
| allSW = rbind(cbind(rep(0,side-1),X[-1,-side]),rep(0,side)) | |
| # summation of the matrices | |
| X2 <- allW + allNW + allN + allNE + allE + allSE + allS + allSW | |
| # the rules of GoL are applied using logical subscripting | |
| X3 <- X | |
| X3[X==0 & X2==3] <- 1 | |
| X3[X==1 & X2<2] <- 0 | |
| X3[X==1 & X2>3] <- 0 | |
| X <- X3 | |
| # each simulation step is stored | |
| storage[,,i] <- X2 | |
| # note that I am storing the array of Ni values - | |
| # - this is in order to make the animation prettier | |
| } | |
| storage <- storage/max(storage) # scaling the results | |
| # to a 0-1 scale | |
| ## ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ | |
| ## End of Original Codes | |
| ## ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ | |
| ## ============================================================================= | |
| ## Create Animated GIF using 'rBlocks' and 'animation' | |
| ## ============================================================================= | |
| ## unique values and colours | |
| value_unique <- sort(unique(matrix(storage, ncol=1)), decreasing = F) | |
| colour_unique <- colorRampPalette(brewer.pal(9,"Greys"))(length(value_unique)+1)[-1] | |
| ## Create GIF (500 x 500) | |
| library(animation) | |
| saveGIF({ | |
| for (n_frame in (skips+1):steps){ ## drop the first n frames (burn out) | |
| ## Extract from storage | |
| a <- storage[, , n_frame] | |
| block_a <- make_block(a) | |
| ## Update Colours | |
| for (n in 1:length(value_unique)) { | |
| block_a[which(a[,] == value_unique[n])] <- colour_unique[n] | |
| } | |
| ## Display | |
| display(block_a) | |
| } | |
| }, movie.name = "game_of_life_arena.gif", interval = 0.1, nmax = steps, ani.width = 500, | |
| ani.height = 500) |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| ## ============================================================================= | |
| ## The #rBlocks Experiement - Pixelation | |
| ## ============================================================================= | |
| ## Load EBImage (install if needed) | |
| if (!suppressMessages(require(EBImage))) { | |
| source("http://bioconductor.org/biocLite.R") | |
| biocLite("EBImage") | |
| } | |
| ## Load packages | |
| suppressMessages(library(RColorBrewer)) | |
| suppressMessages(library(animation)) | |
| suppressMessages(library(rblocks)) | |
| ## Core Parameters | |
| set.seed(1234) # for reproducible results | |
| num_pixel <- 50 # Height and Width (only resize to a square for now) | |
| ## Read and resize image | |
| ## Download this and put it in the same working directory https://dl.dropboxusercontent.com/u/103222/R/R.png | |
| img_raw <- resize(readImage("R.png"), w = num_pixel, h = num_pixel) | |
| ## Convert into Greyscale 256 (0-255) | |
| img_grey <- channel(img_raw, "grey") | |
| img_mat <- t(round((1-imageData(img_grey)) * 255, digits = 0) + 1) | |
| ## need to add 1 for data slicing and then t() to correct rotation | |
| ## Create block template | |
| block_template <- make_block(matrix(1, num_pixel, num_pixel)) | |
| ## Create sets of colours (un-comment to enable more) | |
| set_colour <- list( | |
| #Accent <- colorRampPalette(brewer.pal(8,"Accent"), interpolate = "spline")(256), | |
| #Dark2 <- colorRampPalette(brewer.pal(8,"Dark2"), interpolate = "spline")(256), | |
| #Paired <- colorRampPalette(brewer.pal(12,"Paired"), interpolate = "spline")(256), | |
| #Set1 <- colorRampPalette(brewer.pal(9,"Set1"), interpolate = "spline")(256), | |
| #Set2 <- colorRampPalette(brewer.pal(8,"Set2"), interpolate = "spline")(256), | |
| #Set3 <- colorRampPalette(brewer.pal(12,"Set3"), interpolate = "spline")(256), | |
| #Pastel1 <- colorRampPalette(brewer.pal(9,"Pastel1"), interpolate = "spline")(256), | |
| #Pastel2 <- colorRampPalette(brewer.pal(8,"Pastel2"), interpolate = "spline")(256), | |
| Blues <- colorRampPalette(brewer.pal(9,"Blues"), interpolate = "spline")(256), | |
| BuGn <- colorRampPalette(brewer.pal(9,"BuGn"), interpolate = "spline")(256), | |
| BuPu <- colorRampPalette(brewer.pal(9,"BuPu"), interpolate = "spline")(256), | |
| GnBu <- colorRampPalette(brewer.pal(9,"GnBu"), interpolate = "spline")(256), | |
| Greens <- colorRampPalette(brewer.pal(9,"Greens"), interpolate = "spline")(256), | |
| Greys <- colorRampPalette(brewer.pal(9,"Greys"), interpolate = "spline")(256), | |
| Oranges <- colorRampPalette(brewer.pal(9,"Oranges"), interpolate = "spline")(256), | |
| OrRd <- colorRampPalette(brewer.pal(9,"OrRd"), interpolate = "spline")(256), | |
| PuBu <- colorRampPalette(brewer.pal(9,"PuBu"), interpolate = "spline")(256), | |
| #PuBuGn <- colorRampPalette(brewer.pal(9,"PuBuGn"), interpolate = "spline")(256), | |
| PuRd <- colorRampPalette(brewer.pal(9,"PuRd"), interpolate = "spline")(256), | |
| Purples <- colorRampPalette(brewer.pal(9,"Purples"), interpolate = "spline")(256), | |
| RdPu <- colorRampPalette(brewer.pal(9,"RdPu"), interpolate = "spline")(256), | |
| Reds <- colorRampPalette(brewer.pal(9,"Reds"), interpolate = "spline")(256), | |
| YlGn <- colorRampPalette(brewer.pal(9,"YlGn"), interpolate = "spline")(256)) | |
| #YlGnBu <- colorRampPalette(brewer.pal(9,"YlGnBu"), interpolate = "spline")(256), | |
| #YlOrBr <- colorRampPalette(brewer.pal(9,"YlOrBr"), interpolate = "spline")(256), | |
| #YlOrRd <- colorRampPalette(brewer.pal(9,"YlOrRd"), interpolate = "spline")(256), | |
| #BrBG <- colorRampPalette(brewer.pal(9,"BrBG"), interpolate = "spline")(256), | |
| #PiYG <- colorRampPalette(brewer.pal(9,"PiYG"), interpolate = "spline")(256), | |
| #PRGn <- colorRampPalette(brewer.pal(9,"PRGn"), interpolate = "spline")(256), | |
| #PuOr <- colorRampPalette(brewer.pal(9,"PuOr"), interpolate = "spline")(256), | |
| #RdBu <- colorRampPalette(brewer.pal(9,"RdBu"), interpolate = "spline")(256), | |
| #RdGy <- colorRampPalette(brewer.pal(9,"RdGy"), interpolate = "spline")(256), | |
| #RdYlBu <- colorRampPalette(brewer.pal(9,"RdYlBu"), interpolate = "spline")(256), | |
| #RdYlGn <- colorRampPalette(brewer.pal(9,"RdYlGn"), interpolate = "spline")(256), | |
| #Spectral <- colorRampPalette(brewer.pal(9,"Spectral"), interpolate = "spline")(256)) | |
| ## Identify unique colours | |
| colour_unique <- sort(unique(as.integer(img_mat))) | |
| ## Randomise choices | |
| set_rand <- sample(length(set_colour), length(set_colour)) | |
| ## Animate it | |
| saveGIF({ | |
| for (n_frame in 1:length(set_rand)) { | |
| n_set <- set_rand[n_frame] | |
| use_colour <- unlist(set_colour[n_set]) | |
| block_temp <- block_template | |
| for (n_colour in 1:length(colour_unique)) { | |
| block_temp[which(img_mat == colour_unique[n_colour])] <- use_colour[as.integer(colour_unique[n_colour])] | |
| } | |
| display(block_temp) | |
| } | |
| }, movie.name = "test.gif", interval = 0.25, nmax = length(set_rand), ani.width = 500, | |
| ani.height = 500) |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| ## ============================================================================= | |
| ## The #rBlocks Experiement - Pixelation 2 | |
| ## ============================================================================= | |
| ## Load EBImage (install if needed) | |
| if (!suppressMessages(require(EBImage))) { | |
| source("http://bioconductor.org/biocLite.R") | |
| biocLite("EBImage") | |
| } | |
| ## Load packages | |
| suppressMessages(library(animation)) | |
| suppressMessages(library(rblocks)) | |
| ## Function - pixelate | |
| pixelate <- function(name_input = NULL, | |
| name_output = "output.gif", | |
| pixel_seq = seq(5, 75, 5), | |
| gif_interval = 0.15, | |
| gif_width = 500, | |
| gif_height = 500, | |
| gif_reverse = TRUE) { | |
| ## Read Image | |
| img_raw <- readImage(name_input) | |
| ## Add reversed pixel sequence if needed | |
| if (gif_reverse) { | |
| pixel_rev <- sort(pixel_seq[which(pixel_seq != min(pixel_seq) & | |
| pixel_seq != max(pixel_seq))], | |
| decreasing = T) | |
| pixel_seq <- c(pixel_seq, pixel_rev) | |
| } | |
| ## Resize, pixelate and animate | |
| saveGIF({ | |
| for (n_seq in 1:length(pixel_seq)) { | |
| ## Reisze | |
| img_resized <- resize(img_raw, w = pixel_seq[n_seq], h = pixel_seq[n_seq]) | |
| ## Convert to hex colour and rotate with t() | |
| img_hex <- t(channel(img_resized, "x11")) | |
| ## Create rBlocks | |
| block <- make_block(matrix(NA, pixel_seq[n_seq], pixel_seq[n_seq])) | |
| ## Update Colours | |
| block[,] <- img_hex[,] | |
| ## Display rBlocks for animation | |
| display(block) | |
| } | |
| }, | |
| movie.name = name_output, | |
| interval = gif_interval, | |
| nmax = 100, | |
| ani.width = gif_width, | |
| ani.height = gif_height) | |
| } | |
| ## Example 1 - rCrimemap (https://dl.dropboxusercontent.com/u/103222/R/rcmap.png) | |
| pixelate("rcmap.png") | |
| ## Example 2 - http://rcharts.io/img/slidify_logo_notext.png (converted into jpg without transparent background) | |
| pixelate("slidify_logo_notext.jpg") | |
| pixelate("slidify_logo_notext.jpg", gif_width = 200, gif_height = 200) # smaller version |
Hi Pssguy,
Thanks! I didn't get email notification for gist so I missed your comment until now.
For now, my short answers are:
a) Yes. The EBImage package is more flexible than just a square. It can resize the image to pre-defined height and width.
b) I am also new to the animation package. If I find the answer, I will let you know.
I am busy working on two papers right now but I will get back to this later!
Cheers,
Joe
Hi Pssguy,
Re (a) I just had a quick look at the EBImage::resize(). It is actually super easy to resize while preserving proportion. Just omit either the "h" or "w" parameter and it will do the rest.
Re (b) still no idea why the "loop = FALSE" failed. Which operation system did you run your test on?
Joe
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment






This is wonderful. I am trying to adapt test_04
a) Are there possibilities of resizing other than to a square
b) I would like to run the animation for a while and then stop. I'm unfamiliar with the options
and when trying loop=FALSE got the error
convert.exe: invalid argument for option `-loop': FALSE @ error/convert.c/ConvertImageCommand/1998.