Before you code, do three things:
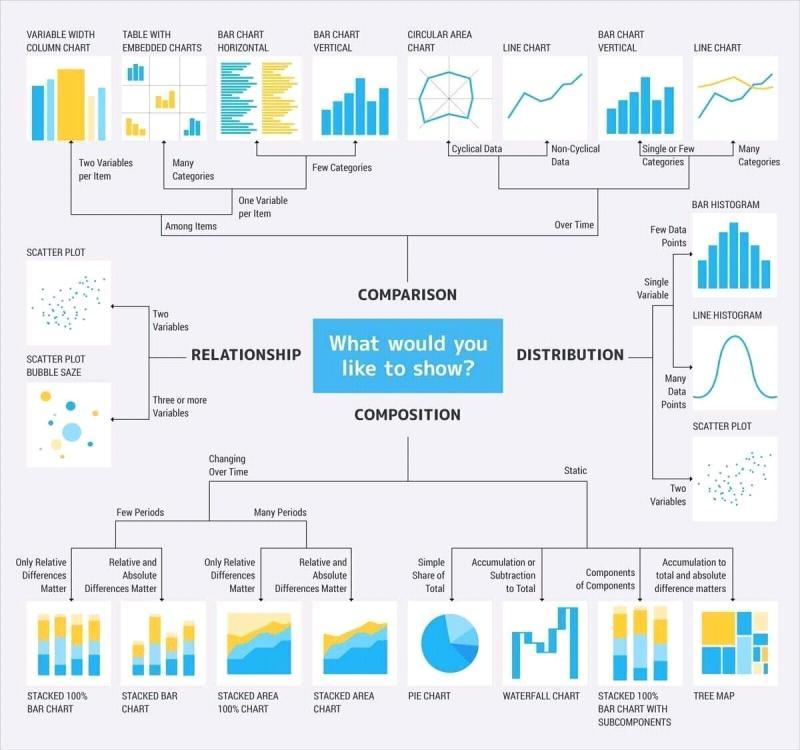
- Think about what you want to show in your data. For example, how would you like to ultimately visualize it or what would you ultimately like to say about it? Here are some helpful info graphics from a data science cheatsheet article on Medium:
-
Do quick initial visualizations of your data to get a sense of their distributions, whether it's actually worth doing a statistical test (i.e., does it look like there might be a significant difference?), and whether there are any outliers (which could be interesting in themselves! E.g., why did that particular participant choose xyz vs. how the rest of the group chose?). Diverging stacked bar charts are excellent as first visualizations for ordinal data (like Likert Scale data), and box plots are also excelent first visualizations for other data. JMP is apparently great for these types of initial quick-looks, and Vega-Lite (e.g., Vega-Lite editor, normalized stacked bar chart) is awesome for creating visualizations right in the browser with JSON (e.g., use csv to json converter or python pandas to_json) or with Python/Jupyter.
- With these plots, you'll get a sense of:
- the distribution (e.g., if it's skewed to the left or right, it might not be normal),
- see whether the two distributions you're comparing are similar (and if not, figure out if there are different subgroups of the data that are causing different distributions),
- see if there are outliers (and why this might be the case → Is there a particular type of person/variable that's causing the outliers?), and
- figure out if it's worth it to do a statistical test (e.g., if the distributions are similar, but the medians are different).
- Here are some useful links about how to analyze box plots:
- Interpreting box plots by Khan Academy (e.g., T/F statements about box plots)
- How to compare box plots by Linh Ngo
- More on how to compare box plots by Linh Ngo
- See this paper about why diverging stacked bar charts are ideal for visualizing Likert scale results.
- With these plots, you'll get a sense of:
-
Figure out which type of statistical test you need to use; for example, by using the flowchart below from this article.

-
The first question to ask yourself is: Is your data normal? In many of the code snippets below, this question is taken care of for you by a normality test, and the appropriate test will automatically be chosen (e.g., if normal --> t-test).
- Note also that if you have a sample size n > 30, "according to the central limit theorem, the distribution of the sample mean satisfies the normal distribution when the number of samples is above 30", so you do not necessarily have to check for normality [Normality Test in Clinical Research, Central limit theorem: the cornerstone of modern statistics].
-
The second question is: Is your data paired or unpaired? Paired data is usually where you follow individuals through the data, like when you're trying to find differences between pre/post surveys. Unpaired data is where you're finding differences between different "groups" of participants, like parents vs. children on a specific survey. For paired data, you use tests like the "paired t-test" (normal) and the "Wilcoxon signed-rank test" (not normal). For unpaired data, you use tests like the "independent t-test" (normal) and the "Mann-Whitney U test" (not normal).
-
Other tests might include Kendall Tau correlation, Krippendorff's alpha for computing inter-coder reliability for thematic analysis, etc.
-
If you're considering doing a lot of comparisons because you have a large number of factors/covariates that could potentially affect your dependent variable, don't do it! Doing many comparisons (e.g., with a t-test) increases the chances of incorrect results. Use an omnibus test, like ANOVA/ANCOVA/MANCOVA/Friedman/etc. instead! Quote from "Biofeedback Heart Rate Training during Exercise" by Goldstein et al.:
The experimental data were analyzed in two steps. First, analyses of
variance (ANOVAs) were performed and then independent means t-tests
comparing the experimental groups. The reason for this apprach was that
the data analysis involved multiple between-group comparisons, and the
probability that at least one of these would reveal a statistically significant
difference by chance alone was unacceptably high. ANOVAs provided an
assessment of the significance of the overall effects of feedback, sessions,
etc., on the dependent variables, and obtaining significant effects in the
ANOVAs permitted application of multiple t-tests to the data without fear
of wrongly inferring significant differences between groups.
Here's a link to a fantastic explanation of the differences between ANOVA, ANCOVA, MANOVA, & MANCOVA. Below, pictures from the site explain the differences in a nutshell.
Side Note:
- "Dependent Variable" == output (e.g., someone's heart rate)
- "Independent Variables" == input variables that don't change (e.g., users' age or the app version), which can be either "factors" or "covariates"
- "Covariates" == continuous independent variable (e.g., age, number of hours spent studying, or anxiety-trait score)
- "Factor" == independent variable that comes in "groups" and is not continuous (e.g., gender, level of education, or app version)
- "Omnibus Test" == finds whether there is an overall difference across groups, before doing post-hoc tests—like t-tests—which can find the source of the difference (more info)
- "Repeated Measures" == when you measure the same thing at different times (e.g. blood pressure of a group of people at three different times), or the same thing with different 'treatments' (e.g. engagement of a group of people with three different apps), you can used Repeated Measures tests, like Repeated Measures ANOVA, ANCOVA, Friedman test, etc. (more info 1, more info 2)
Side Note:
ANOVA, ANCOVA, linear regression, etc. methods expect the output ("dependent") variable to be continuous. If you have oridinal data, like Likert scale data, technically you're supposed to use ordinal methods instead, like ordered logistic regression, multinomial logistic regression, or binary logistic regression [Analysing Likert Scale Type Data]. That being said, you can treat Likert scale data as continuous if either:
- (1) the Likert scale has five or more categories (Johnson & Creech, 1983; Norman, 2010; Sullivan & Artino, 2013; Zumbo & Zimmerman, 1993), or
- (2) you can average two or more ordinal variables (e.g., Likert scale questions) to create an approximately continuous variable.
| One-Way ANOVA | Two-Way ANOVA |
|---|---|
 |
 |
- Input: one or two factors (i.e., with levels/groups)
- Output: one continuous variable
 (Basically ANOVA, but also includes at least one covariate / continuous independent variable.)
(Basically ANOVA, but also includes at least one covariate / continuous independent variable.)
- Input: factors and covariates
- Output: one continuous variable
Here are some links describing ANCOVA and Linear Regression [ANCOVA expressed as linear regression, How to perform ANCOA in Python, How to fit models using formulas & categorical variables]. Essentially, you can use the statsmodels.api package, statsmodels.formula.api package, and the ols function to model your data (Linear Regression), and get summary statistics. Use the p-values in the summary to determine if they likely affect the outcome of the dependent variable.
Make sure to test the assumptions for linear regression though! See this post on how to test linear regression assumptions in Python.
Here's a code snippet of how to model data with both categorical (C()) and covariate independent variables.
import pandas as pd
import statsmodels.api as sm
import statsmodels.formula.api as smf
# Example dataframe:
df = pd.read_csv("avg-hr-by-app-type.csv")
df.head()| avgHR | appType | expLevel | gender | anxietyTrait | age | busyness |
|---|---|---|---|---|---|---|
| 57.956522 | A | advanced | male | 1.6 | 19 | 0.5025 |
| 88.900000 | B | novice | female | 1.6 | 31 | 0.4775 |
| 100.388889 | A | advanced | male | 2.0 | 42 | 0.4775 |
| 105.981132 | B | novice | female | 1.4 | 22 | 0.3450 |
| 82.645161 | A | advanced | male | 1.0 | 39 | 0.3600 |
# ANCOVA (Note: C --> categorical variable)
formula = "avgHR ~ C(appType) + C(expLevel) + C(gender) + anxietyTrait + age + busyness"
model = smf.ols(formula=formula, data=df).fit()
model.summary()At the top of the model summary, find the R-squared and the Prob (F-statistic) values. These tell you the goodness-of-fit. See Regression Analysis by Example by Chatterjee & Hadi for in-depth discussion of linear regression.
E.g., "The goodness-of-fit index, R^2, may be interpreted as the proportion [percent] of the total variability in the response variable Y that is accounted for by the predictor variable[s]"
E.g., "H_0 is rejected if ... p(F) <= alpha", and "H_0: Reduced model is adequate ... H_1: Full model is adequate" (i.e., if p < alpha, the full model/formula you developed is likely significant!)
If you can't satisfy the linearity assumption or can't come up with a model where p < alpha, R^2 is great enough, etc., consider adding more variables to your formula and/or interaction effects, or transforming the dependent variable (e.g., log(y)). Here's some more info on transformations:
- The best explanation I've found for why we're allowed to use these transformations is that the transformation functions preserve the order of the output—they're monotonic transformations
- Detailed examples of transformations (log, Box-Cox): Statistical Methods II: Data Transformations
- Packages in R for transformations (note the 'log', 'Tukey’s Ladder of Powers', 'Box-Cox' transformations): R Handbook: Transforming Data
- Often-cited explanation of transformations: Transformation and Weighting in Regression by Carrol & Ruppert)
| One-Way MANOVA | Two-Way MANOVA |
|---|---|
 |
 |
(Basically ANOVA, but has multiple output/dependent variables.)
- Input: one or two factors
- Output: multiple continuous variables
 (Basically ANCOVA, but has multiple output/dependent variables.)
(Basically ANCOVA, but has multiple output/dependent variables.)
- Input: factors and covariates
- Output: multiple continuous variables
See the following code snippets for how to do stats in Python:
-
Paired t-test, Wilcoxon, and other useful helper functions
- E.g., Did students trust conversational agents more on the pre-test than on the post-test?
-
Unpaired t-test, Mann-Whitney, and other useful helper functions
- E.g., Did parents trust conversational agents less than children?
-
Kendall Tau correlation matrix and test
- E.g., Is trust correlated with friendliness of the agent?
-
Krippendorff's alpha with MASI distance for intercoder reliability
- E.g., How much did Researcher A agree with Researcher B when tagging data?
-
Clean text for word cloud and create word cloud
- E.g., Did students mention "technology" more than the word "computer" in their long-answer questions?
-
Variable association with Fisher's exact test
- E.g., Was there an association between [children and commenting on human-likeness of an agent] and [parents and commenting on artificiality of an agent]?
More detail about methods used above: