— by Tobias Kuhn, 4 April 2022; reporting on work with Cristina Bucur, Davide Ceolin, and Jacco van Ossenbruggen [1,2] —
I believe we have made the first steps venturing into a new era of scientific publishing. Let me explain. [Update: At what point exactly a new era begins and what counts as first steps are of course subject to debate. I therefore added a section on related initiatives and further reading below.]
Science is nowadays communicated in a digital manner through the internet. We essentially have a kind of "scientific knowledge cloud", where researchers with the help of publishers upload their latest findings in the form of scientific articles, and where everybody who is interested can access and retrieve these findings. (This is in fact only true for articles that are published as Open Access, but that is not the point here.)
In a sense, this scientific knowledge cloud has been a big success, but it also has serious limitations. Imagine somebody wanting to build an application that feeds on all these findings, for example to help researchers learn about interesting new developments or to summarize scientific consensus for laypeople. Concretely, a developer might face a task like this one:
Retrieve all genes that have been found to play a role in a part of the respiratory system in Covid-19 patients. Only include results from randomized controlled trials published in the last seven days.
To a naive developer without experience in how scientific knowledge is communicated, this might sound quite easy. One would just have to find the right API, translate the task description into a query, and possibly do some post-processing and filtering. But everybody who knows a bit how science is communicated immediately realizes that this will take a much bigger effort.
The problem is that scientific results are represented and published in plain text, and not in a structured format that software could understand. So, in order to access the scientific findings and their contexts, one has to apply text mining first. Unfortunately, despite all the impressive progress with Deep Learning and related techniques in the past few years, text mining is not good enough, and probably will never be.
To illustrate the point, we can look at the results of the seventh BioCreative workshop held in November 2021, where world-leading research teams competed in extracting entities and relations from scientific texts. Just to detect the type of a relation between a given drug and a given gene out of 13 given relation types, the best system achieved an F-score of 0.7973.
But that is just the relation type. To get the full relation out, we first have to know the entity on the left-hand side (subject) and right-hand side (object) of the relation. We can look at a different task of the BioCreative workshop to get a feeling of how well extracting these subjects and objects work. The task focussed on extracting chemicals, and this is done it a two-stage process. First the entities are recognized in the text, with an F-score of the best system of 0.8672, and then the recognized chemicals are linked to the corresponding formal identifiers, with the best F-score being 0.8136:
A very rough back-of-the-envelope calculation can give us an estimate of the quality of mining such entire relations:
An overall F-score of 0.40, as resulting from this calculation, roughly means that around 60% of retrieved relations are wrong and 60% of existing relations are not retrieved. This is clearly not even close to good enough for most types of possible applications. And mind you, these numbers come from the best performing systems when world-leading research groups compete and probably put months of effort into optimizing their systems for the specific problem. Moreover, we are talking here only about the simplest possible kind of relations of the form subject-relation-object.
This seems to point to a deeper problem. Text mining is just a work-around, and the real problem is elsewhere. As Barend Mons rhetorically asked "Why bury it first and then mine it again?". Instead of seeing text mining as the ultimate solution, we should just stop burying our knowledge in the first place.
In the work I will explain below, we wanted to find out how we can practically publish findings without burying them.
As a first step to experiment with such a new way of publishing, we needed to find a general way of how to represent high-level scientific findings in some sort of formal logic. Even though such findings are arguably the most important outcome of science, there was no prior work on practically mapping (most of) these findings to formal logic across domains. To better understand the logical structure of such high-level scientific findings (e.g. that a gene tends to have a certain effect on the course of a given disease), we selected a random sample of 75 research articles from Semantic Scholar.
Studying the high-level findings from these random articles, we managed to elicit a wide-spread logical pattern, which we then turned in the super-pattern model. It consists of five slots to be filled, and translates directly to a logical formula. Our paper [1] explains the details, and I give here just one example. The finding of the article entitled "P‐glycoprotein expression in rat brain endothelial cells: evidence for regulation by transient oxidative stress" can be expressed with the super-pattern as follows:
- Context class: rat brain endothelial cell
- Subject class: transient oxidative stress
- Qualifier: generally
- Relation: affects
- Object class: Pgp expression
The three class slots take any class identifier (here shown by class name for simplicity), whereas the qualifier and the relation come from closed lists of a few possible values. Informally, the example above means that whenever there is an instance of "transient oxidative stress" in the context of an instance of "rat brain endothelial cell" then generally (being defined as in at least 90% of the cases) it has a relation of type "affects" to an instance of "Pgp expression" that is in the same context. Formally, it corresponds to this logical formula (in a bit of a non-standard notation using conditional probability):
Once we discovered this pattern we tried to formalize the 75 high-level findings, and this was the result:
68 of the 75 findings, shown in blue, could be represented with the super-pattern. And the remaining 7 findings, shown in red, are in fact easier to represent, as they can be captured with a simple subject-relation-object structure. (So these latter simpler structures are the ones that existing text mining approaches struggle with, and it is safe to assume that they would perform even worse on the more complicated statements here shown in blue.)
So, it seems that we have found a logical pattern that allows us to represent most high-level scientific findings from different disciplines.
Next, we wanted to make a serious practical step into the new era where findings are machine-interpretable from the start. We designed this as a kind of field study with a special issue of machine-interpretable papers at an existing journal. We wanted these papers to look like regular papers to those who want to look at them in that way, but they should also come with representations in formal logic for anyone or anything that knows how to deal with that. For that special issue, we chose the journal Data Science, of which I am an editor-in-chief.
We also had to make a practical concession though: While the whole setup could be used to publish novel findings, we restricted ourselves to findings from existing publications. For that, we introduced the concept of a formalization paper whose novel contribution is the formalization of an existing finding. So, authors of a formalization paper take credit for the formalization of the finding, but not for the finding itself.
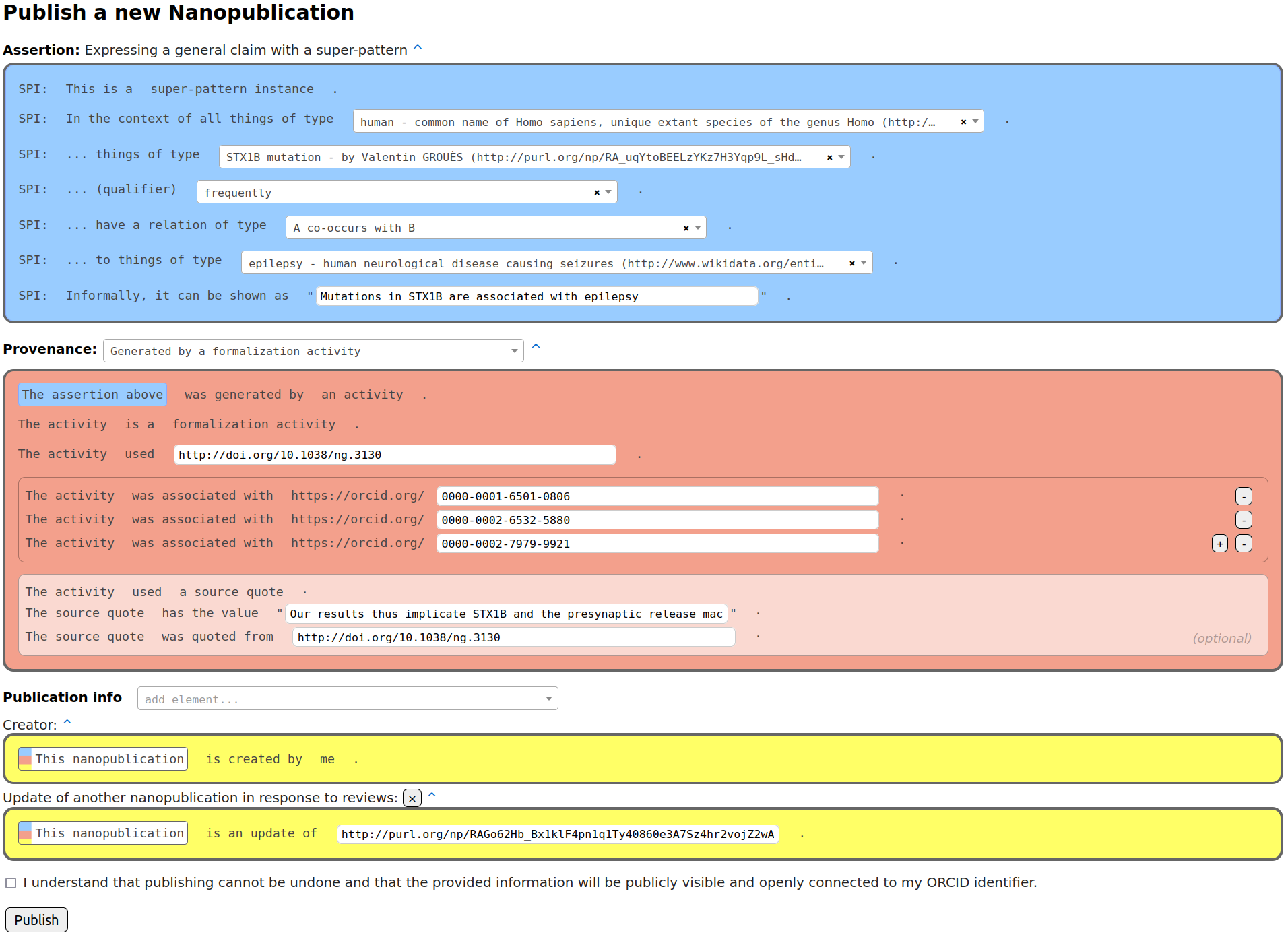
To represent these findings and thereby the formalization papers, we used the nanopublication format and the Nanobench tool. Researchers who contributed to this special issue filled in a form to express and submit their formalization that looked like this:
Apart from the actual formalization in the assertion part of the nanopublication (blue), this also involved specifying the provenance of that formalization (red) and further metadata (yellow). The provenance in this case states that the assertion was derived by a formalization activity taking an existing paper as input. This interface provides auto-completing drop-down menus and maps the result to the logic language RDF in the back. I won't go into the details of defining new classes and reviewing (both done with nanopublications and Nanobench too), but you can have a look at the preprint of our paper [2] on that.
We ended up with 15 formalization papers in our special issue, as summarized by this table:
Each row of this table corresponds to a formalization, and can be written out as a logic formula. So, these papers look here quite different from what we are used to. But if you go to the official page for the special issue on the publisher's website, they look like regular papers. Besides the regular button to download the paper as a PDF, there is also a link that points to the nanopublication representation, thereby connecting the two ways of looking at this paper:
So, for the first time, software can reliably interpret the main high-level findings of scientific publications. This special issue is a just a small first step, but it could prove to be the first step into a new era of scientific publishing. The practical immediate consequences of this might seem limited, but I think the longer-term potential of making scientific knowledge accessible to the interpretation by machines is hard to overstate.
- Cristina-Iulia Bucur, Tobias Kuhn, Davide Ceolin, Jacco van Ossenbruggen. Expressing high-level scientific claims with formal semantics. In Proceedings of K-CAP '21. ACM, 2021.
- Cristina-Iulia Bucur, Tobias Kuhn, Davide Ceolin, Jacco van Ossenbruggen. Nanopublication-Based Semantic Publishing and Reviewing: A Field Study with Formalization Papers. arXiv 2203.01608, 2022.
For completeness, I list here some related initiatives and further reading. I only include references where machine-interpretable representations of findings are part of or treated as scientific publications.
- Initiative: GloBI with its integrations to Plazi/Zenodo and Pensoft
- Journal: Biodiversity Data Journal
- Viewpoint paper: Nathan S Upham, Jorrit H Poelen, Deborah Paul, et al. Liberating host–virus knowledge from biological dark data. The Lancet Planetary Health 5.10, 2021
- Implementation: Lyubomir Penev, Terence Catapano, Donat Agosti, Teodor Georgiev, Guido Sautter, Pavel Stoev. Implementation of TaxPub, an NLM DTD extension for domain-specific markup in taxonomy, from the experience of a biodiversity publisher, In Journal Article Tag Suite Conference (JATS-Con) Proceedings 2012
- Editorial: Lyubomir Penev, David Roberts, Vincent Smith, Donat Agosti, Terry Erwin. Taxonomy shifts up a gear: New publishing tools to accelerate biodiversity research. ZooKeys 50, 1-4. 2010
- Position paper: Tobias Kuhn and Michel Dumontier. Genuine semantic publishing. Data Science, 1(1-2), 2017
You can write a comment below to suggest additions to this list.
This text is available under the CC BY 4.0 license. The images were created with Excalidraw, using several of its libraries of visual elements, and are available under the MIT license.







Very nice to hear about this work @tkuhn ! Yes, as @jhpoelen mentioned, our Biodiversity Knowledge Hub taskforce has been thinking a lot about the creation of semantic knowledge graphs for ecological interaction data -- specifically as relates to mammals, viruses, and our evidence of their interactions (including type of assay used, etc). We summarized some of these points last year in this Lancet Planetary Health article, including as relates to the extraction of old data (Plazi), new ways of publishing (Pensoft), and how to connect all these data (GloBI): https://www.thelancet.com/journals/lanplh/article/PIIS2542-5196(21)00196-0/fulltext
It would be great to connect our group with yours to think about this more.
Cheers, --nate